timeline
title R Workshop Series 2025
May 14-15 : Beginner Workshop (2 days)
Sep 18-19 : Intermediate Workshop (2 days)
Nov 20-21 : Advanced Workshop (2 days)
R Beginners Course 2025
Introduction to R and Basic Programming Concepts
Bioinformatics Core Facility CECAD
2025-05-13
Welcome to the R Workshop Series!

- Learning R step-by-step
Welcome to the R Workshop Series!

- Learning R step-by-step
- Focus: Molecular Biology and Data Analysis
Welcome to the R Workshop Series!

- Learning R step-by-step
- Focus: Molecular Biology and Data Analysis
- Building reproducible research skills (at data level)
Workshop Goals

Five Goals
Why Learn R as a Biologist?
- Handle biological data (e.g., gene expression, sequencing)
Why Learn R as a Biologist?
- Handle biological data (e.g., gene expression, sequencing)
- Access and use bioinformatics packages (Bioconductor!)
Why Learn R as a Biologist?
- Handle biological data (e.g., gene expression, sequencing)
- Access and use bioinformatics packages (Bioconductor!)
- Automate and streamline analyses
Why Learn R as a Biologist?
- Handle biological data (e.g., gene expression, sequencing)
- Access and use bioinformatics packages (Bioconductor!)
- Automate and streamline analyses
- Reproducibility and transparency
Why Learn R as a Biologist?
- Handle biological data (e.g., gene expression, sequencing)
- Access and use bioinformatics packages (Bioconductor!)
- Automate and streamline analyses
- Reproducibility and transparency
- Create publication-ready graphs
Workshop Schedule
- Beginner Workshop: May
- Intermediate Workshop: September
- Advanced Workshop: October
Practice between workshops is key!
Timeline
Learning Between Workshops

Learning Between Workshops
- Learning R = Learning a language: Use it often!

Learning Between Workshops
- Learning R = Learning a language: Use it often!
- Practice exercises, small projects

Learning Between Workshops
- Learning R = Learning a language: Use it often!
- Practice exercises, small projects
- Apply R to your own data

Learning Between Workshops
- Learning R = Learning a language: Use it often!
- Practice exercises, small projects
- Apply R to your own data
- Prepare questions for next workshop

What You’ll Learn in This Workshop (revisted)
- Basics of R and RStudio
- Variables, data types, and structures
- Data input/output and reshaping
- Basic plotting with base R
- control flows and apply functions
- Introduction to reproducible reports (R Markdown)
🛠 Focus: Base R only — no extra packages yet!
Why Start with Base R?
- Learn R’s core concepts: data types, operations, programming basics
Why Start with Base R?
- Learn R’s core concepts: data types, operations, programming basics
- Build strong foundations for any R analysis
Why Start with Base R?
- Learn R’s core concepts: data types, operations, programming basics
- Build strong foundations for any R analysis
- Understand how R handles data internally
Why Start with Base R?
- Learn R’s core concepts: data types, operations, programming basics
- Build strong foundations for any R analysis
- Understand how R handles data internally
- Prepare for more advanced workflows later
Why Start with Base R?
- Learn R’s core concepts: data types, operations, programming basics
- Build strong foundations for any R analysis
- Understand how R handles data internally
- Prepare for more advanced workflows later
In future workshops, we will explore modern tools like the tidyverse — but good base R skills come first!
Tools We’ll Use
- R (The software)
- RStudio (Integrated development environment)
- R Markdown (Literate programming)
- Base R functions and structures (The language)
Structure of Each Workshop
- Lectures combined with live demonstrations
- Hands-on exercises
- Mini projects
- Open Q&A sessions
Learning by doing!
Workshop Norms
- No bad questions
- Practice actively — typing > watching
- Help/encourage each other
- Respect different learning paces
The Journey Ahead
- Beginner: Base R Foundations
- Intermediate: Advanced Concepts, Tools and Applications
- Advanced: Reproducible Research and Bioinformatics
Step-by-step towards good data analysis skills!
Let’s Get Started!
🎯 Setting up RStudio and exploring R basics…
Slides & Code

- [f] Full screen
- [o] Slide Overview
- [c] Notes
- [h] help
git repo
Clone repo
git clone https://github.com/CECADBioinformaticsCoreFacility/Beginners_R_Course_2025.git
Slides Directly
https://cecadbioinformaticscorefacility.github.io/Beginners_R_Course_2025/
Session 1 :: Background & R Basics
The Lifeline of R
The Lifeline of R
- R was born as an implementation of S in 1993, because
native S had gone commercial - Being free and encouraging contributions by the user community,
R can easily "evolve" to adapt to new needs and trends Data driven science, including the genome projects, was the perfect “niche” which R could successfully claim for itself- Indeed the
Bioconductor projectwas initiated by one of the founders of R
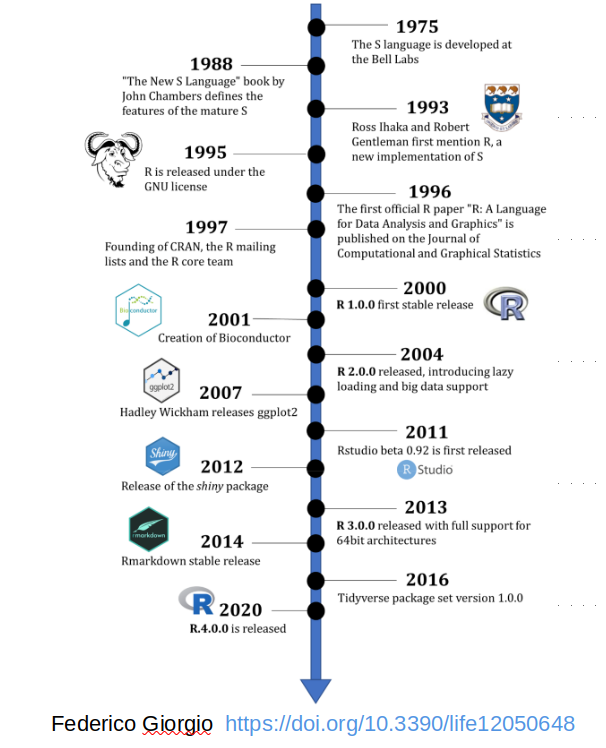
The Lifeline of R
- The
RStudio (now: Posit)company is gaining increasing influence on the evolution of the language- its
Integrated Development Environment (IDE)is increasingly used by people doing data analysis with R - Posit is actively developing and promoting the
tidyverse, which is both a special style and a code repository for the analysis ofdata tables
- its
- Currently there is quite some
evolutionary pressurefor change of the language!

The Iris Data of Edgar Anderson and Ronald A. Fisher
– Some Background Information on Our Practice Dataset
We will use the iris dataset of floral traits for practicing throughout the course:
The Iris Data of Edgar Anderson and Ronald A. Fisher
– Some Background Information on Our Practice Dataset
- Data published by Ronald A. Fisher in 1936
- Statistician and co-founder of population genetics
- Plants collected by Edgar Anderson
- I. setosa and I. versicolor in 1935,
in the same natural habitat - I. virginica likely in 1926,
at a different place
- I. setosa and I. versicolor in 1935,
- Field botanist with a focus on speciation mechanisms
The Iris Data of Edgar Anderson and Ronald A. Fisher
– Some Background Information on Our Practice Dataset
Both men were involved in the making of the Modern/Evolutionary Synthesis, with complementary central tenets:
- R. A. Fisher: Model evolutionary processes from the known facts of genetics
- E. Anderson: Observe the real dynamics of (plant) populations, in order to understand the role of genetics in evolution
The Iris Data of Edgar Anderson and Ronald A. Fisher
– Some Background Information on Our Practice Dataset
Edgar Anderson suspected that I. versicolor may be an allopolyploid hybrid:
I. versicolor =
I. setosa (2n) x I. virginica (4n)
(confirmed by Lim et al. 2007)This may have supported the establishment of the species, by preventing back-crossing to its parents.
Fisher applied his Linear Discriminant Analysis technique to Anderson’s data, in order to test the hypothesis of additive gene action:
if true, versicolor should be twice as similar to virginica than to sertosa!
Interacting R
Session 2 :: Basic Concepts in R
Variables
- Variables are containers for storing data values.
- R does not have a command for declaring a variable
- A variable is created the moment you first assign a value to it.
- Assignment operator
<-/=can be used for assigning a value
Variable Names
A variable can have a short name (like x and y) or a more descriptive name (age, carname, total_volume). Rules for R variables are:
- A variable name must start with a letter and can be a combination of letters, digits, period(.) and underscore(_).
Data Types
| Data Type | Example | Verify | value |
|---|---|---|---|
| Logical | TRUE / FLASE | x<-TRUE print(x) class(x) |
TRUE logical |
| Numeric | 1.3, 5, 4.2 | x<-1.35 print(x) class(x) |
1.35 numeric |
| Integer | 1L, 0L, 4L | x<-35L print(x) class(x) |
35 integer |
| Complex | 2+3i | x<-2+3i print(x) class(x) |
2+3i complex |
| Character | “Hello!” | x<-"Hello!" print(x) class(x) |
Hello! character |
R Data Structure
The variables are assigned with R-Objects and the data type of the R-object becomes the data type of the variable. There are many types of R-objects. The frequently used ones are −
[1] "100" "200" "450" "670"Operators
Operators are the symbols that tell the compiler to perform specific mathematical or logical manipulations. R language is rich in built-in operators and provides the following types of operators −
| Operator | Name | Example |
|---|---|---|
| + | Addition | x + y |
| - | Subtraction | x - y |
| * | Multiplication | x * y |
| / | Division | x / y |
| ^ | Exponent | x ^ y |
| %% | Modulus (Remainder from division) | x %% y |
| %/% | Integer Division | x%/%y |
| Operator | Name | Example |
|---|---|---|
| == | Equal | x == y |
| != | Not equal | x != y |
| > | Greater than | x > y |
| < | Less than | x < y |
| >= | Greater than or equal to | x >= y |
| <= | Less than or equal to | x <= y |
| Operator | Description |
|---|---|
| & | Element-wise Logical AND operator. It returns TRUE if both elements are TRUE |
| && | Logical AND operator - Returns TRUE if both statements are TRUE |
| | | Elementwise- Logical OR operator. It returns TRUE if one of the statement is TRUE |
| || | Logical OR operator. It returns TRUE if one of the statement is TRUE. |
| ! | Logical NOT - returns FALSE if statement is TRUE |
| Operator | Description | Example |
|---|---|---|
| : | Creates a series of numbers in a sequence | x <- 1:10 |
| %in% | Find out if an element belongs to a vector | x %in% y |
| %*% | Matrix Multiplication | x <- Matrix1 %*% Matrix2 |
Session 3 :: Data I/O and Reshaping
Data IO (read & write files)
In this session, we’ll try build an understanding of base R functions for data input/output (I/O) and data reshaping using the
irisdataset.Beyond simply running code, we’ll discuss why you might choose one function over another, highlighting their specific strengths and trade-offs, when it makes sense.
We’ll to this in an interactive way.
CSV formats
Base R provides a versatile suite of I/O functions. Some are highly configurable (e.g., read.table()), while others wrap common defaults for convenience (e.g., read.csv()).
write.csv(iris, "iris.csv", row.names = FALSE)
iris_csv <- read.csv("iris.csv")
head(iris_csv, n = 6) Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5.0 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosawrite.csv2(iris, "iris2.csv", row.names = FALSE)
iris_csv2 <- read.csv2("iris2.csv")
head(iris_csv, n = 6) Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5.0 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosaGeneral Tabular I/O
When you need full control—different delimiters, quoting rules, or no headers— the generic read.table() and write.table() shine. They accept parameters like sep, quote, na.strings, and more.
write.table(iris, "iris_tab.tsv", sep = "\t", row.names = FALSE)
iris_tab <- read.table("iris_tab.tsv", header = TRUE, sep = "\t")
head(iris_tab, n = 10) Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5.0 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa
7 4.6 3.4 1.4 0.3 setosa
8 5.0 3.4 1.5 0.2 setosa
9 4.4 2.9 1.4 0.2 setosa
10 4.9 3.1 1.5 0.1 setosadelim1 <- read.delim("iris_tab.tsv")
# change comma character in data.frame delim1 to comma
delim1$Sepal.Length <- gsub(replacement = ",", pattern="\\.", delim1$Sepal.Length)
delim1$Sepal.Width <- gsub(replacement = ",", pattern="\\.", delim1$Sepal.Width)
delim1$Petal.Length <- gsub(replacement = ",", pattern="\\.", delim1$Petal.Length)
delim1$Petal.Width <- gsub(replacement = ",", pattern="\\.", delim1$Petal.Width)
write.table(delim1, "iris_tab2.txt", sep = "\t", row.names = FALSE)
delim2 <- read.delim2("iris_tab2.txt")
head(delim1, n = 2) Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5,1 3,5 1,4 0,2 setosa
2 4,9 3 1,4 0,2 setosa Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosaR’s Internal Binary Formats
When performance, fidelity and reproducibility matter—especially for large objects—R’s binary formats (.RDS / .RData) beat text. saveRDS() / readRDS() handle single objects, while save() / load() manage multiple objects.
Data Reshaping: Base R Tools
Base R reshaping functions cover pivoting, stacking, grouping, and merging. We’ll compare each pair to understand when to use one versus another.
reshape()handles complex wide⇄long pivots in one call via thedirectionargument. It’s powerful but can be verbose when specifyingvaryingandtimes.stack()/unstack()provide a quick way to collapse or expand multiple columns into key-value pairs, but without the record-level identifiers thatreshape()preserves.
stack() / unstack()
Execute the code. Examine:
- Are values grouped by column (Sepal.Length then Sepal.Width, etc.)?
- How does the ind factor map each measurement?
set.seed(123)
stacked2 <- stacked[sample(nrow(stacked)), ]
unstacked2 <- try(unstack(stacked2), silent = TRUE)
unstacked2 Sepal.Length Sepal.Width Petal.Length Petal.Width
1 7.7 3.4 5.1 0.1
2 4.3 3.8 4.7 1.4
3 5.5 3.4 1.4 1.5
4 5.0 2.9 4.6 2.2
5 5.8 2.3 6.0 1.0
6 4.6 2.0 4.9 0.2
7 6.1 2.8 1.4 2.3
8 6.1 4.4 6.1 1.7
9 6.7 3.0 1.5 0.4
10 5.0 3.1 5.8 2.1
11 5.5 3.2 1.5 1.4
12 6.4 3.2 4.9 1.5
13 4.4 2.8 4.2 0.1
14 5.5 2.5 1.6 0.4
15 4.8 2.9 1.5 0.2
16 6.2 2.9 4.4 1.5
17 5.6 3.0 1.6 1.5
18 6.9 3.1 1.6 2.3
19 7.2 3.0 5.0 0.3
20 6.1 3.4 1.3 0.2
21 5.6 4.0 4.5 2.1
22 5.4 3.5 1.6 1.3
23 7.0 3.4 6.4 0.5
24 6.1 3.0 3.9 0.2
25 6.2 2.6 1.5 0.2
26 6.8 3.5 4.7 1.2
27 5.0 3.1 1.2 0.2
28 6.5 3.6 4.8 1.0
29 6.3 2.8 4.6 0.2
30 4.6 3.0 1.4 0.2
31 6.8 3.1 3.6 2.3
32 5.8 2.3 5.8 1.3
33 5.0 2.7 4.9 1.0
34 5.1 3.0 5.6 1.3
35 6.4 4.2 4.3 1.5
36 5.1 3.0 3.8 1.4
37 4.5 2.5 1.7 0.2
38 6.0 3.5 4.5 2.5
39 5.4 3.0 5.5 1.5
40 4.9 3.4 5.1 2.3
41 5.7 3.5 4.0 0.2
42 5.2 3.2 5.1 1.8
43 5.1 2.5 1.4 0.2
44 4.9 3.3 5.4 1.3
45 6.7 2.8 4.7 1.4
46 5.0 3.1 4.4 2.0
47 5.5 2.8 1.6 1.5
48 5.8 3.0 4.5 2.0
49 4.8 3.2 5.6 0.4
50 6.3 3.0 1.3 1.5
51 6.5 3.0 5.8 1.3
52 6.3 3.0 5.7 1.6
53 6.4 3.1 3.0 2.0
54 7.6 3.0 4.7 0.2
55 5.0 2.8 6.7 0.2
56 4.6 3.0 5.9 0.3
57 6.7 3.8 5.2 2.3
58 6.5 2.9 5.1 1.1
59 6.7 2.8 4.8 1.4
60 4.6 2.2 4.9 0.2
61 6.5 2.7 1.0 2.1
62 7.7 2.6 5.1 1.2
63 5.1 2.7 1.3 0.2
64 6.7 3.1 3.9 1.5
65 6.3 2.8 5.0 0.6
66 5.1 3.0 5.6 1.2
67 7.2 3.7 5.1 2.3
68 6.6 2.9 1.3 0.1
69 5.7 3.8 1.4 1.6
70 5.0 3.7 4.1 1.7
71 6.5 3.0 1.5 0.3
72 6.3 2.7 4.0 1.0
73 5.8 2.9 5.0 1.8
74 4.7 3.5 4.2 1.6
75 5.7 2.6 5.7 1.5
76 5.5 2.7 1.5 0.4
77 4.8 3.3 1.4 1.8
78 4.8 3.4 5.9 0.1
79 5.0 3.7 1.4 1.1
80 5.6 2.5 3.5 1.8
81 5.7 3.1 6.9 1.8
82 7.3 3.4 1.4 2.1
83 5.0 2.6 6.1 1.3
84 5.7 3.2 1.5 0.3
85 5.2 2.4 1.4 1.5
86 5.6 3.0 1.7 1.3
87 7.7 2.7 5.2 1.8
88 5.4 3.8 1.5 2.2
89 6.3 2.3 5.1 1.8
90 7.7 2.5 5.0 1.3
91 6.0 2.9 5.6 0.2
92 7.9 3.0 3.3 1.2
93 5.1 2.7 3.9 1.3
94 5.5 3.0 4.8 0.2
95 6.8 2.8 1.3 0.2
96 5.7 2.2 5.5 2.2
97 7.1 3.9 5.5 2.0
98 6.0 3.2 3.7 0.3
99 4.4 2.5 6.0 1.3
100 7.2 3.2 4.5 0.2
101 4.9 3.3 5.1 1.8
102 6.0 3.2 4.5 1.9
103 6.7 3.9 1.9 1.5
104 6.4 3.6 1.6 0.2
105 5.4 3.3 1.5 2.3
106 6.7 2.2 1.4 1.8
107 6.6 2.8 4.0 1.4
108 6.1 3.4 1.4 2.1
109 5.4 3.2 3.3 1.4
110 5.3 2.8 1.7 1.8
111 6.4 3.0 4.0 0.2
112 7.4 2.4 1.5 0.2
113 5.6 3.0 1.4 0.2
114 6.0 3.4 4.4 2.3
115 4.8 2.7 4.9 1.9
116 5.2 3.0 1.6 0.2
117 6.3 2.7 6.3 0.3
118 5.4 2.8 1.3 2.5
119 6.0 2.5 3.5 0.3
120 6.3 3.2 4.0 1.9
121 6.7 3.0 1.3 0.2
122 5.2 3.6 1.1 2.0
123 6.9 3.1 6.7 1.3
124 5.1 2.5 5.4 2.1
125 6.4 3.3 5.6 0.4
126 5.9 3.1 4.5 0.2
127 5.1 3.1 4.5 1.3
128 6.2 3.5 4.7 1.8
129 4.9 2.8 1.2 1.3
130 5.6 3.4 4.5 1.0
131 5.8 3.2 5.3 1.9
132 6.3 3.6 5.7 1.8
133 6.1 2.8 5.6 0.2
134 5.8 3.2 4.4 1.1
135 4.9 3.4 4.1 2.0
136 6.4 2.9 1.5 1.6
137 4.7 2.3 1.7 2.4
138 5.0 3.2 5.3 1.9
139 4.4 2.6 1.5 1.2
140 6.9 3.0 4.6 1.0
141 6.9 2.4 6.6 2.5
142 5.7 3.0 4.2 2.4
143 5.9 3.3 4.2 0.2
144 5.9 3.4 1.4 0.4
145 5.1 3.0 4.1 2.4
146 6.2 2.9 4.8 0.4
147 5.8 3.8 4.3 1.0
148 4.9 3.8 1.5 1.4
149 5.5 2.9 1.9 0.2
150 5.7 4.1 6.1 0.1reshape()
long <- reshape(
iris,
varying = list(names(iris)[1:4]),
v.names = "Measurement",
timevar = "Feature",
times = names(iris)[1:4],
idvar = c("rowID","Species"),
direction = "long"
)
head(long, 5) Species rowID Feature Measurement
1.setosa.Sepal.Length setosa 1 Sepal.Length 5.1
2.setosa.Sepal.Length setosa 2 Sepal.Length 4.9
3.setosa.Sepal.Length setosa 3 Sepal.Length 4.7
4.setosa.Sepal.Length setosa 4 Sepal.Length 4.6
5.setosa.Sepal.Length setosa 5 Sepal.Length 5.0Summary of Best Practice
- Implicit vs. Explicit IDs: Why do
stack()/unstack()fail after reordering? How doesidvarrescue us? - When to Use Which:
- Quick two-column views →
stack() - Reliable round-trips with extra columns →
reshape()+idvar
- Quick two-column views →
cbind() / rbind() vs. merge()
# Combine first four columns back-to-back plus Species
cb <- cbind(
iris[, 1:2],
iris[, 3:4],
Species = iris$Species
)
head(cb) Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5.0 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa[1] FALSE- Prediction: Does
rb==iris? Not quite: rbind()appends rows but preserves original row names, soidentical(rb, iris)is FALSE. Userownames(rb) <- NULLto align.
- ❌ Error:
cbind()expects equal row counts. It can’t recycle or drop.
Relational Binding: merge()
# Partial views
dir1 <- iris[1:100, c("Sepal.Length","Species")]
dir2 <- iris[51:150, c("Sepal.Width","Species")]
merged <- merge(dir1, dir2, by = "Species")
head(merged, 10) Species Sepal.Length Sepal.Width
1 versicolor 7 2.7
2 versicolor 7 2.0
3 versicolor 7 3.2
4 versicolor 7 3.2
5 versicolor 7 3.1
6 versicolor 7 2.3
7 versicolor 7 2.8
8 versicolor 7 2.8
9 versicolor 7 3.3
10 versicolor 7 2.4- Result: Many-to-many join: e.g. 25
versicolorin each → 625 rows. - Why?
merge()pairs every matching row for duplicated keys.
- Outcome: Exactly one-to-one pairing, recovering the intended alignment.
Try It: Shuffle rows of
dir2before merging—do you still get correct matches? Why? Challenge: Perform a full outer join (all=TRUE) on(Species, idx)and inspect NAs.
4. Summary & Takeaways
cbind()/rbind(): Great for straightforward stacking when dimensions align exactly; no key matching.merge(): Key-based alignment; duplicates produce Cartesian products unless you add an ID for one-to-one matching.
By consciously adding IDs when joining on duplicated keys, you ensure your merged table mirrors your intended relational structure—no surprises!
Other interesting functions
- split() / unsplit() break data into subsets by factor, letting you apply arbitrary functions via lapply() [TOMORROW!], then reassemble.
- table() quickly tabulates counts across factors.
- cut() bins continuous variables into discrete intervals.
Session 4 :: Visualization
R Plots (base) – some preparations
First we will pre-compute the mean values of each flower trait in each species for later use.
R Plots (base) – some preparations
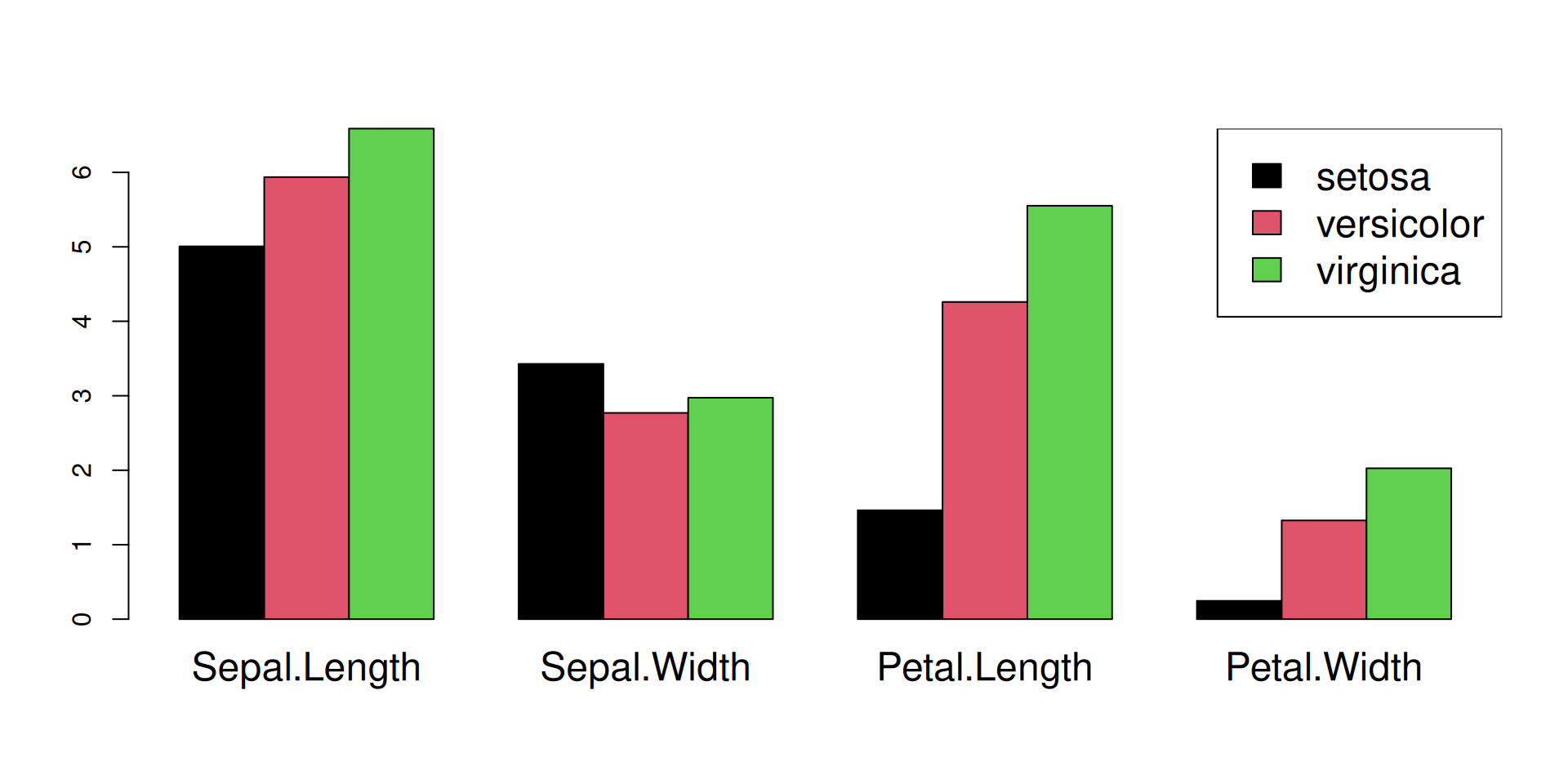
Step 2: Compute the means and assemble them into a matrix
Sepal.Length Sepal.Width Petal.Length Petal.Width
setosa 5.006 3.428 1.462 0.246
versicolor 5.936 2.770 4.260 1.326
virginica 6.588 2.974 5.552 2.026
There are easier ways to run a function over the columns of a table – tomorrow!
R Plots (base) – some preparations
Finally we define our own coloring scheme:
R Plots (base)
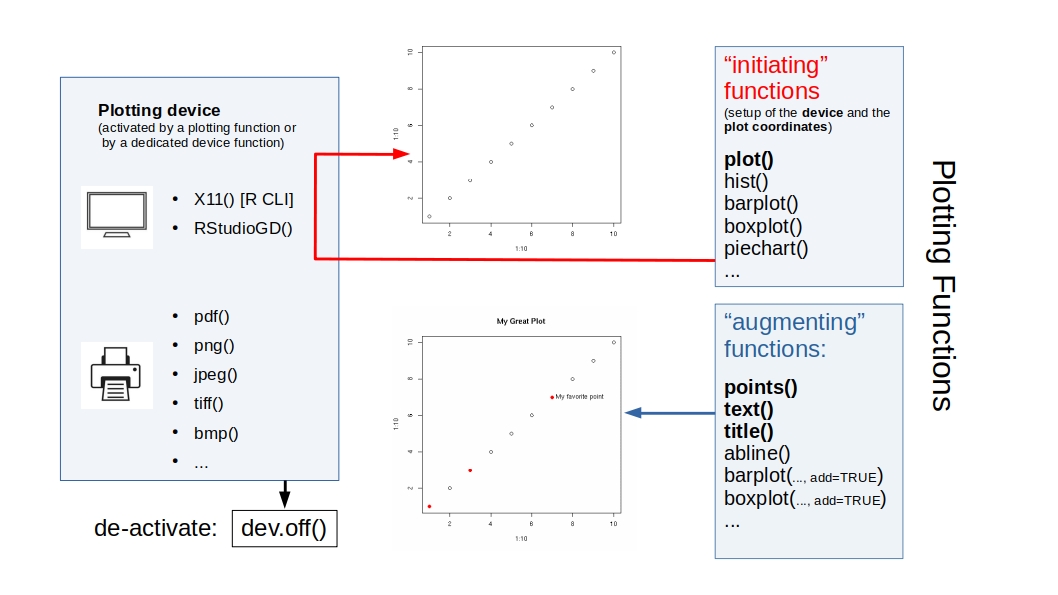
R Plots (base) – barplot()
Barplots represent sign and absolute value of numbers by the direction and length of bars.
If called with a matrix as first argument, the function produces one plot for each column:
R Plots (base) – barplot()
If we want to plot trait means per species, we must change the rows of matrix species_means (= the species) into columns, because barplot() reads a matrix by column.
This is done by the t() function ("transpose"):
m <- t(species_means) ## TRANSPOSE
barplot(## one plot per column == species,
## one bar == trait mean!
m,
## do not stack the bars
beside=TRUE,
## larger group labels
cex.names=2,
col=trait_colors,
## increase y limit to fit the legend
ylim = c(0,10),
cex = 2
)
## add a legend (plot "augmentation"!)
legend(x=1,y=10, ##"topright",
rownames(m),
fill=trait_colors)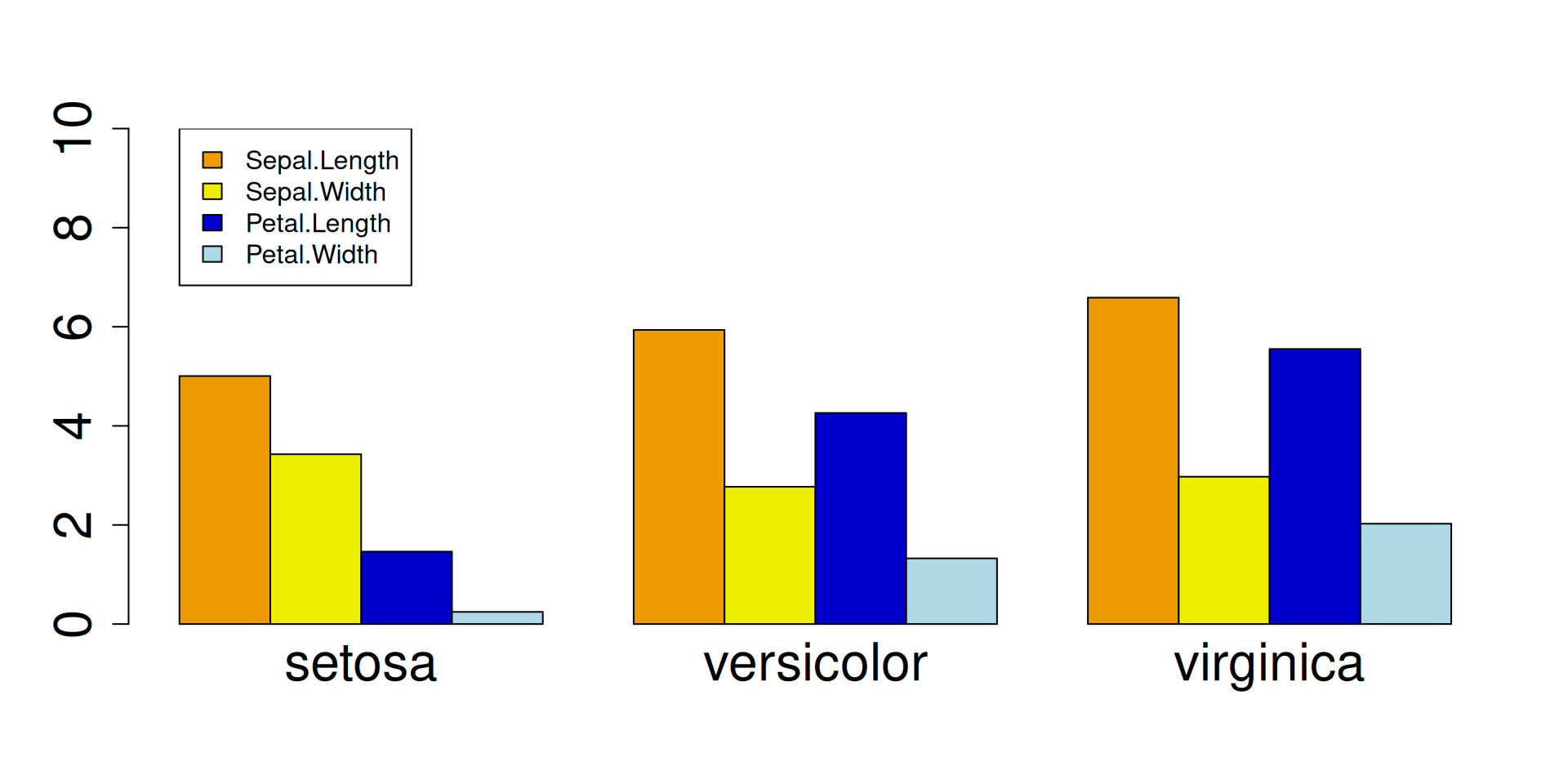
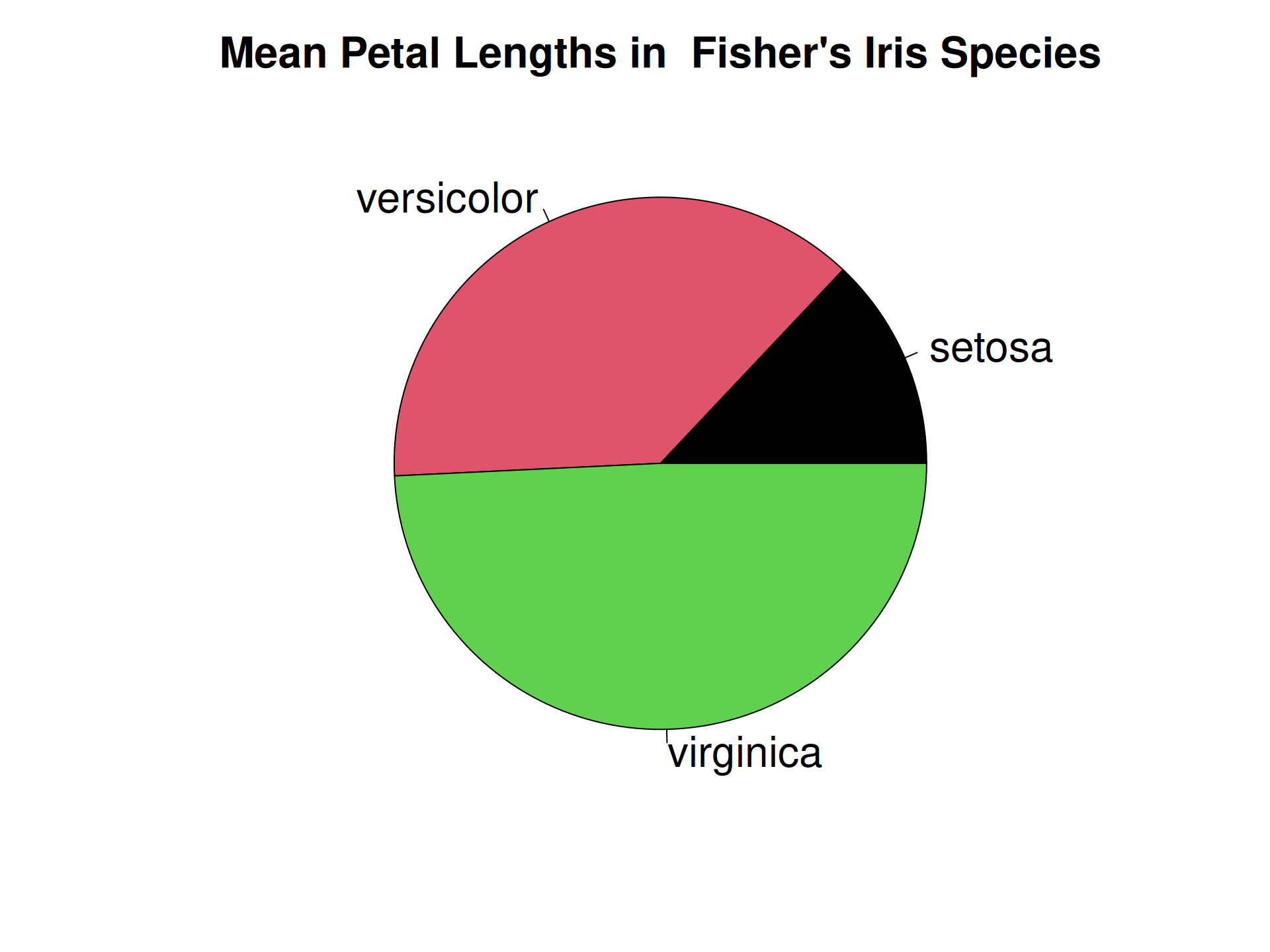
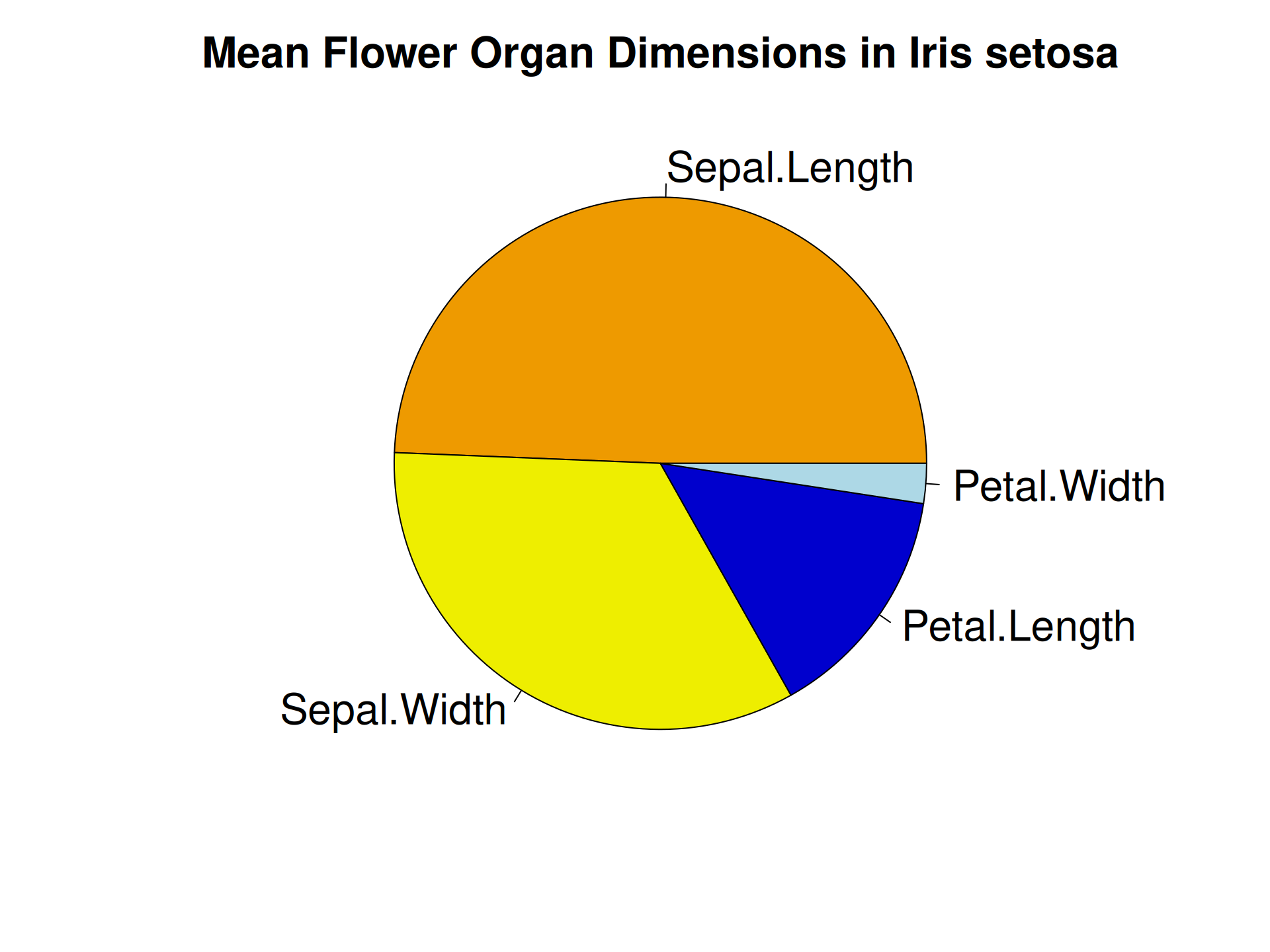
R Plots (base) – pie()
Piecharts are a quick-and-dirty alternative for representing numbers.
The pie() function can only represent one set of numbers at a time. In addition, comparing angles on a piechart is visually not as easy as comparing bar heights.
R Plots (base) – pie()
Piecharts are a quick-and-dirty alternative for representing numbers.
The pie() function can only represent one set of numbers at a time. In addition, comparing angles on a piechart is visually not as easy as comparing bar heights.
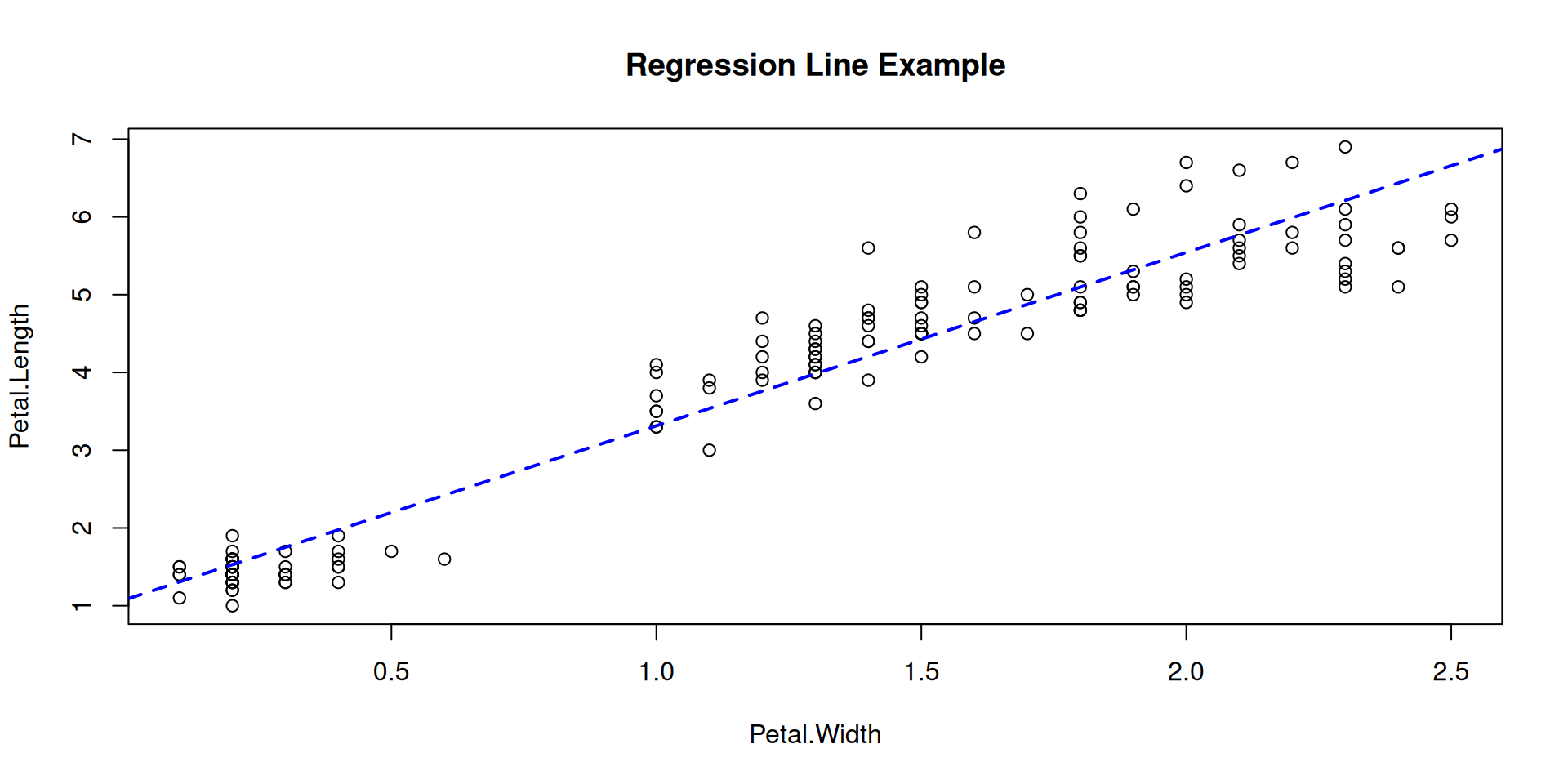
R Plots (base) – plot()
The plot() function is an extremely versatile workhorse for x/y plots.
As an “initializing” function, it may be called to just create an empty canvas, to be filled later:
R Plots (base) – plot()
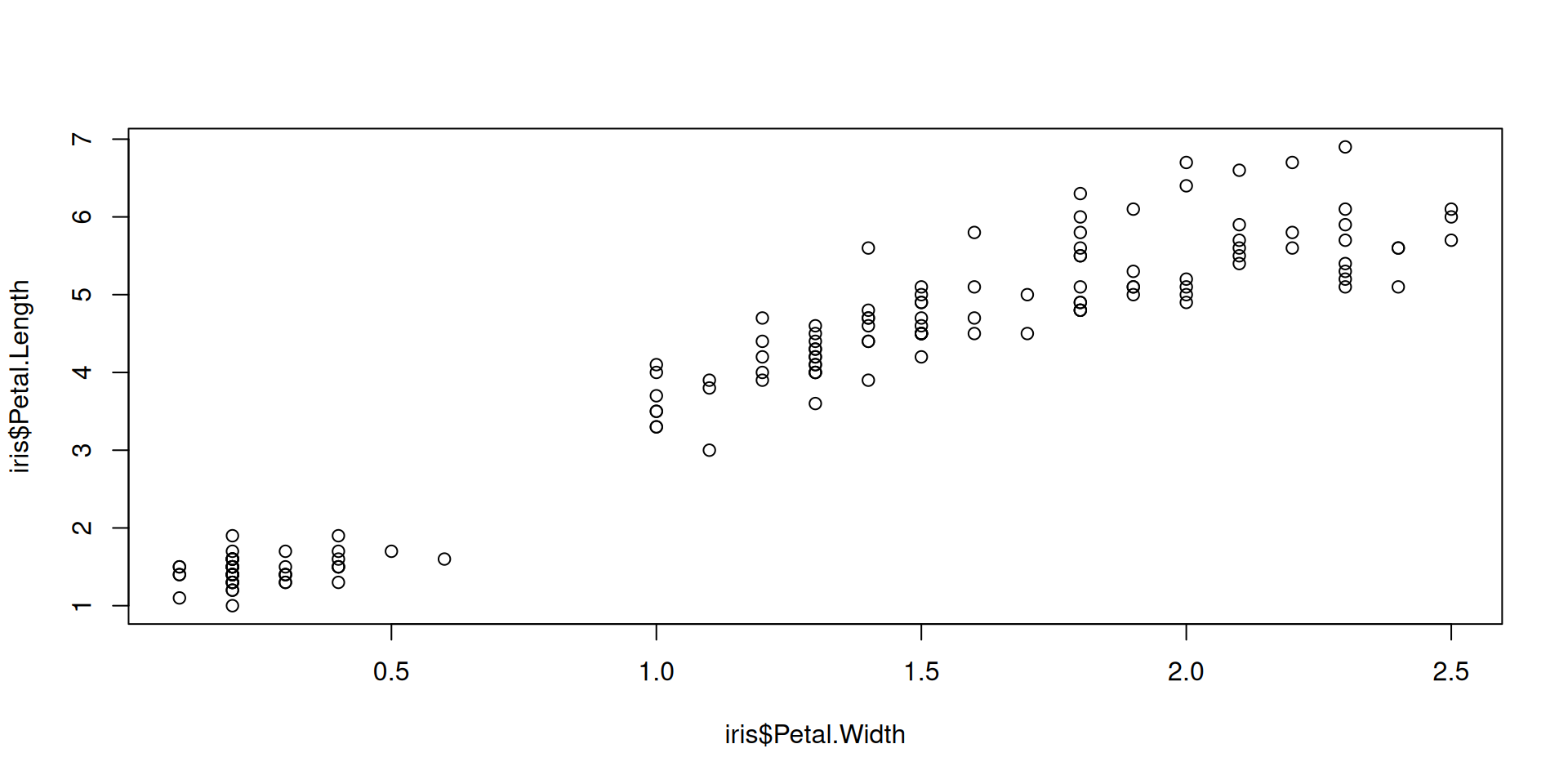
Or it is called with an initial set of data, with the option to extend the plot later:
R Plots (base) – plot()
R Plots (base) – plot()
R Plots (base) – plot()
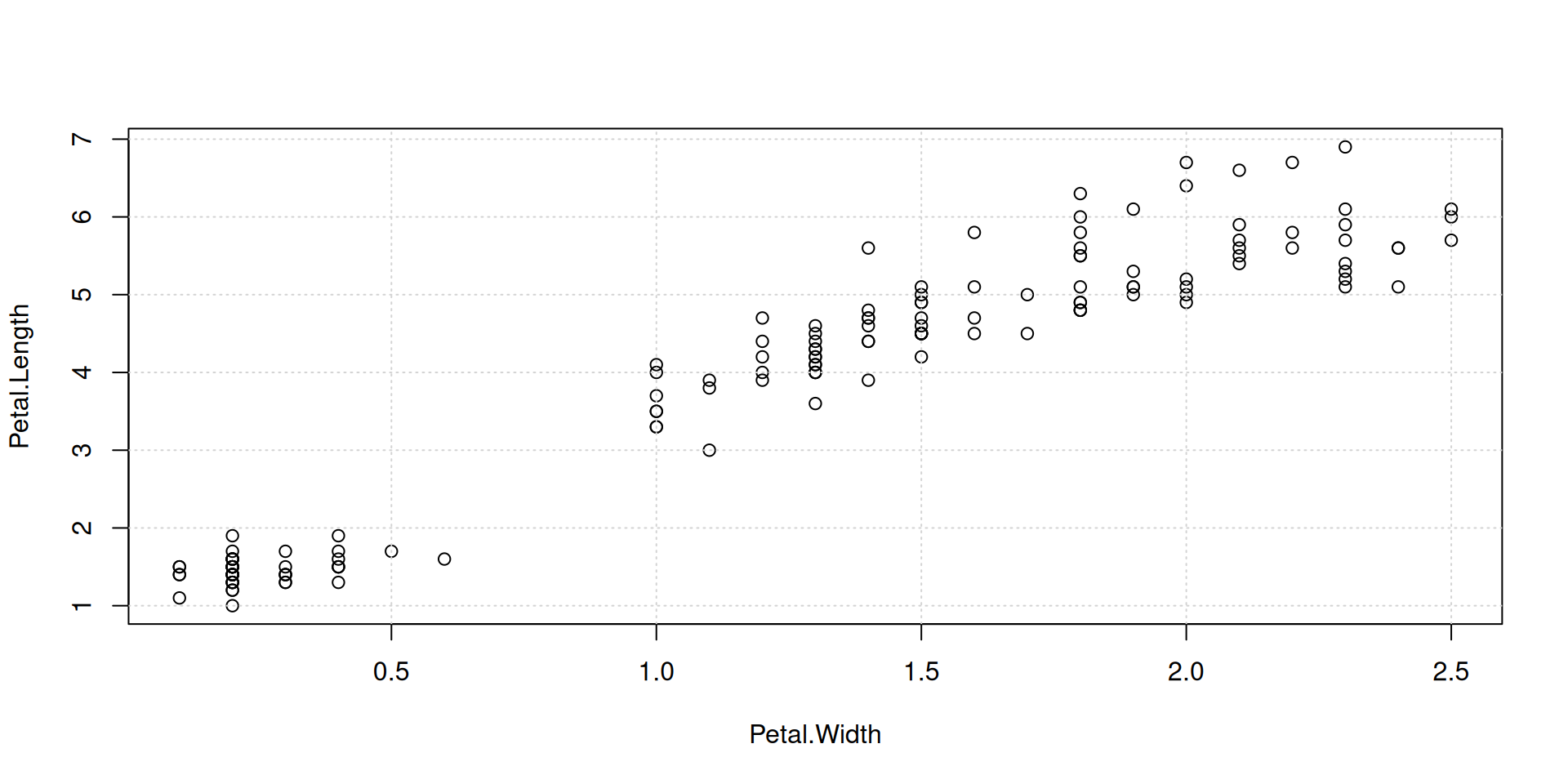
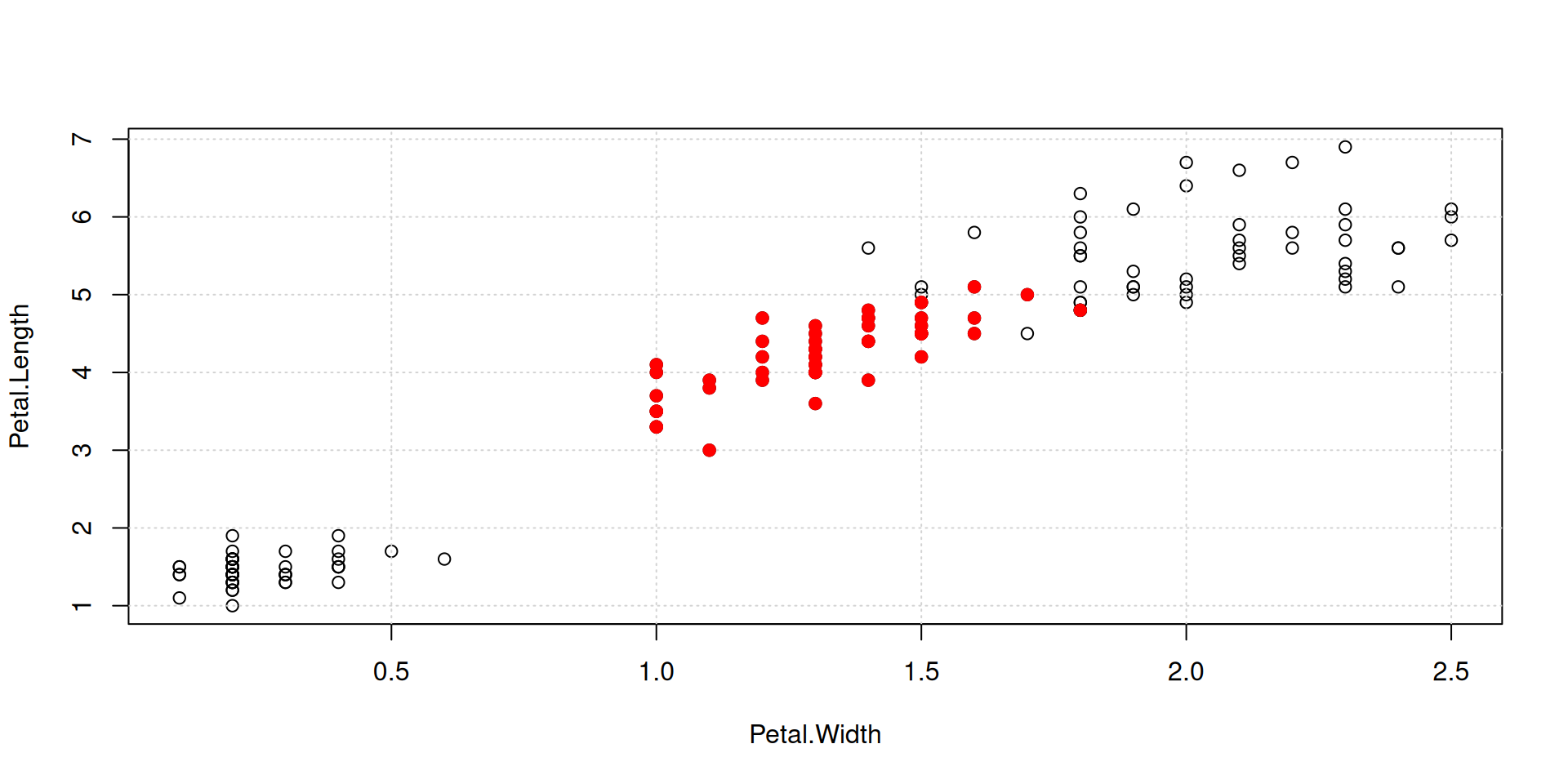
Overplot some points with color, in order to identify a group in your data:
R Plots (base) – plot()
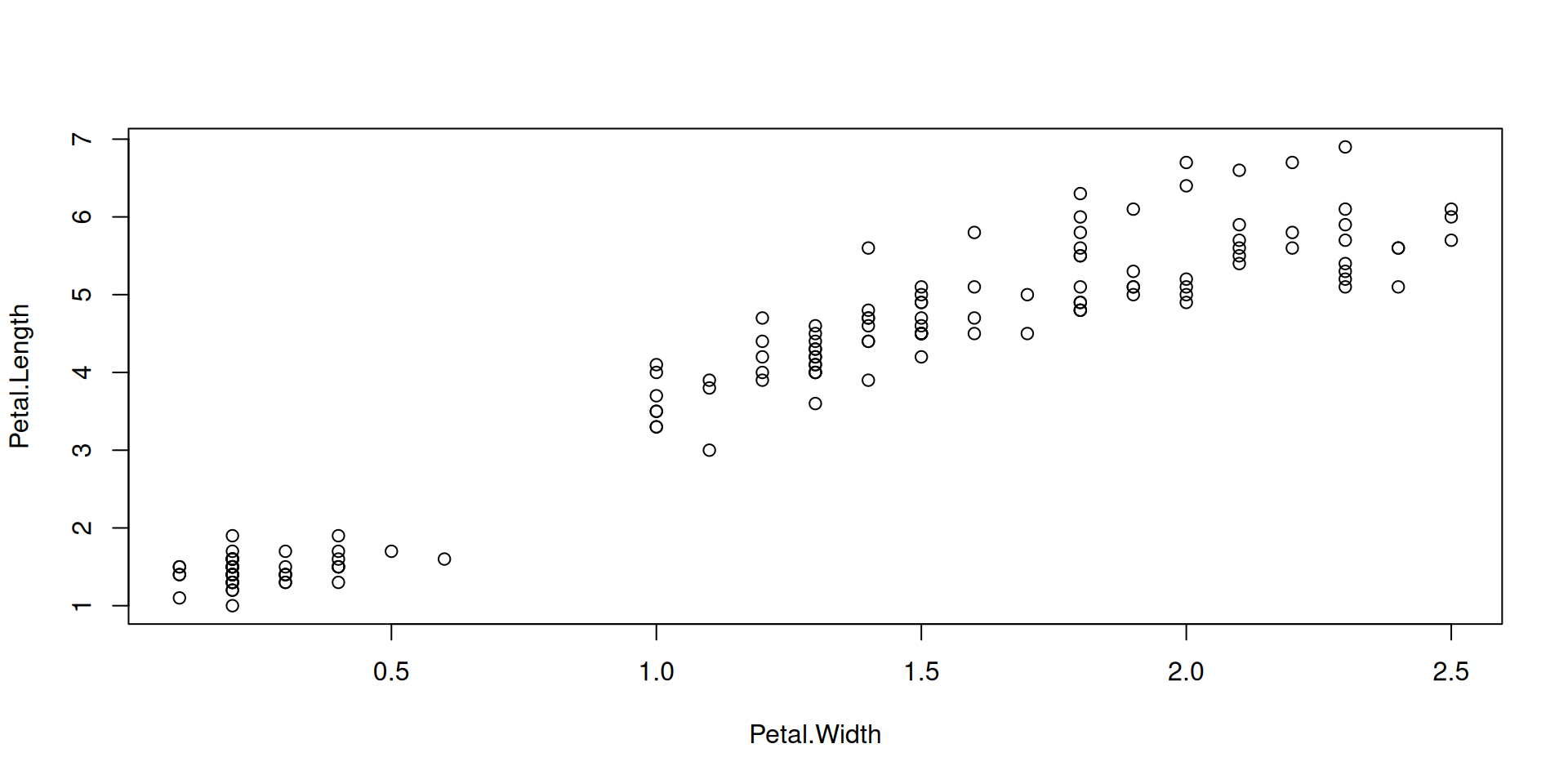
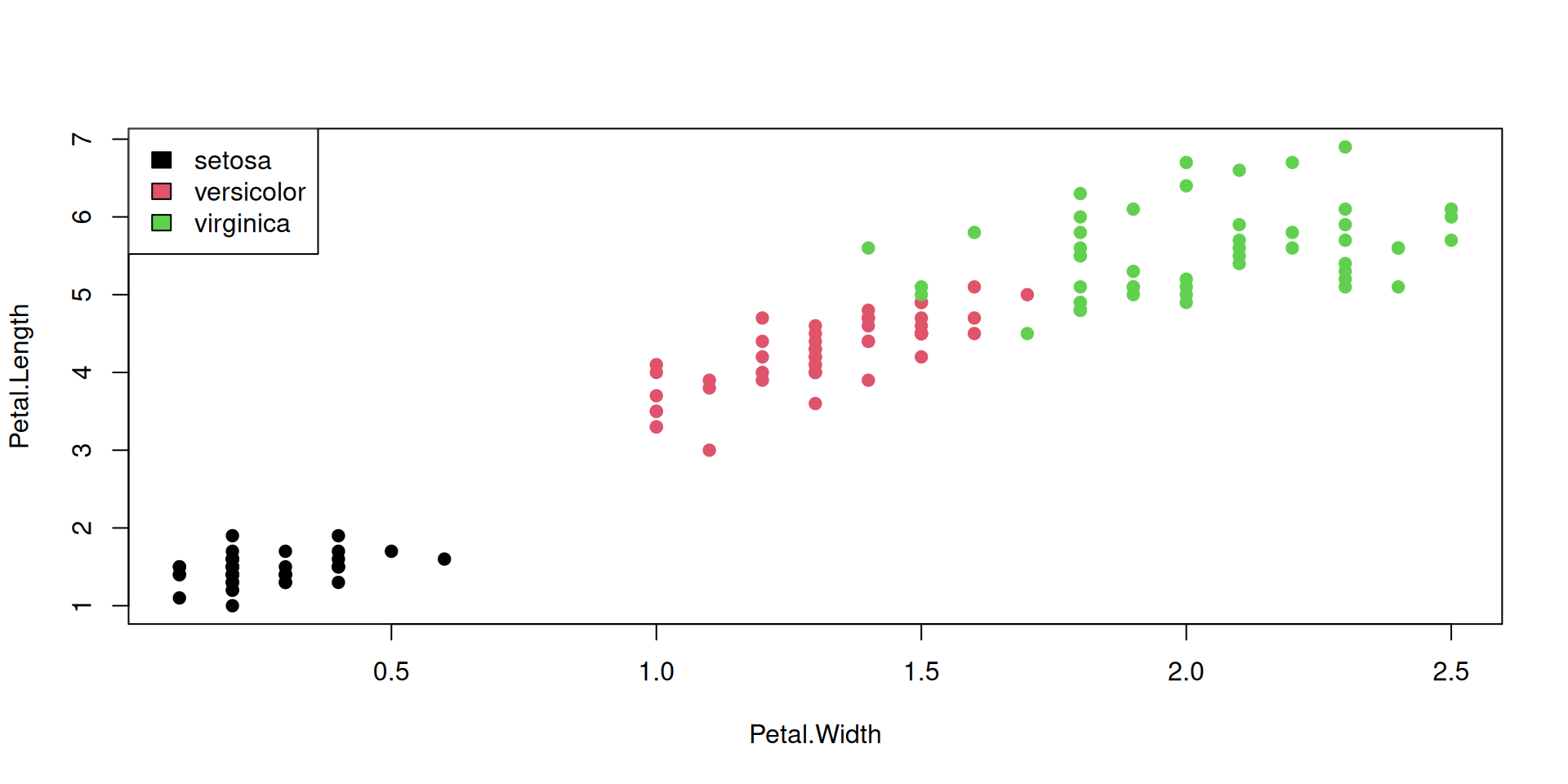
Color all points by species, using our named vector species_colors:
R Plots (base) – plot()
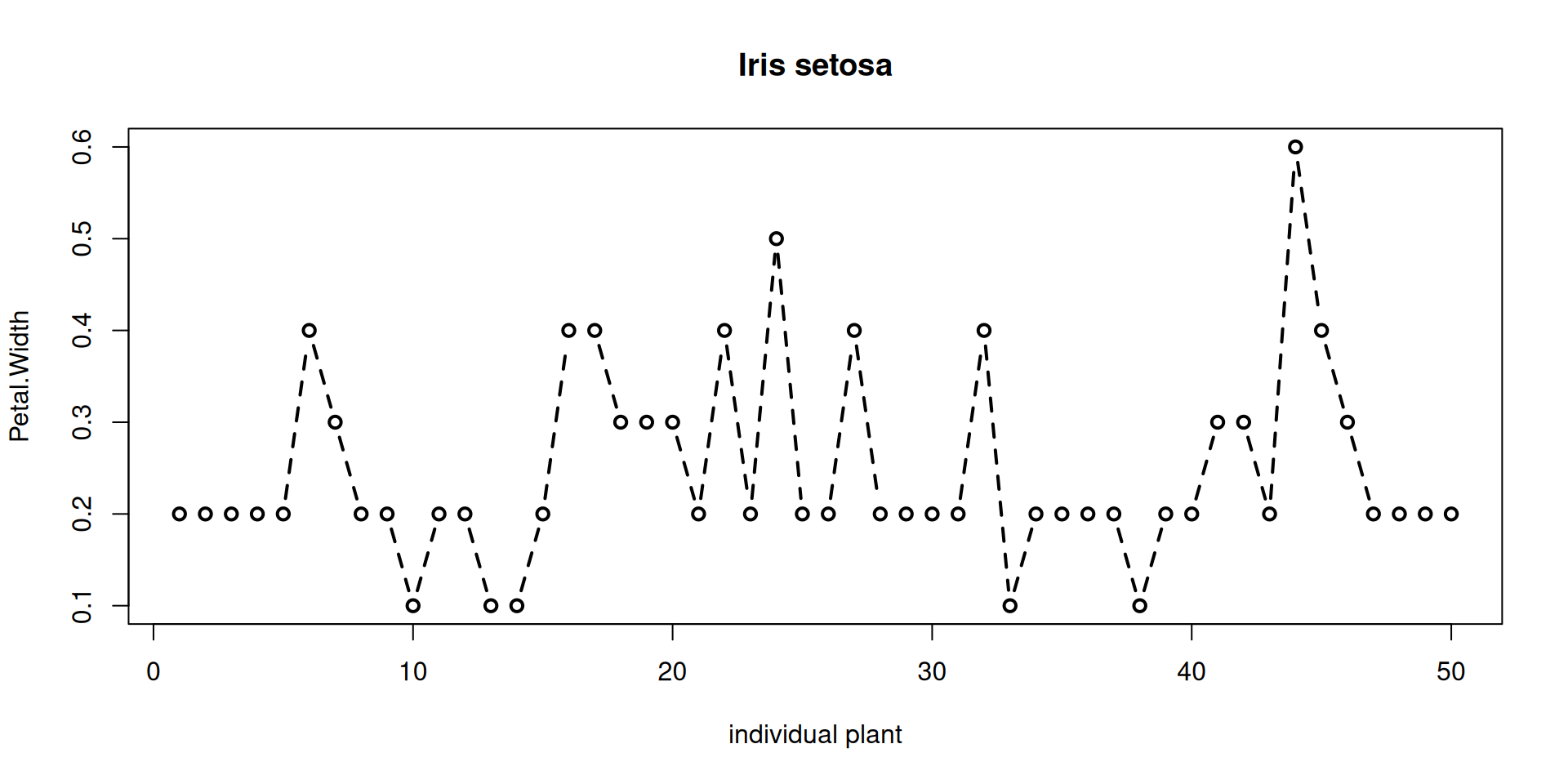
Points with adjacent positions in the input can be connected by lines, using different line styles. A typical use case is a line graph, with x as a running number or ID.
## See par() for line-related parameters!
## Make a new data.frame,
## containing only setosa:
df <- subset(iris, Species=="setosa")
plot(
# x is now the row number in df
x=1:nrow(df),
xlab="individual plant",
y=df$Petal.Width,
ylab="Petal.Width",
## show both points and
## connecting lines:
type="b",
## line width:
lwd = 2,
## line style = dashed:
lty=2,
main="Iris setosa"
)
R Plots (base) – plot()
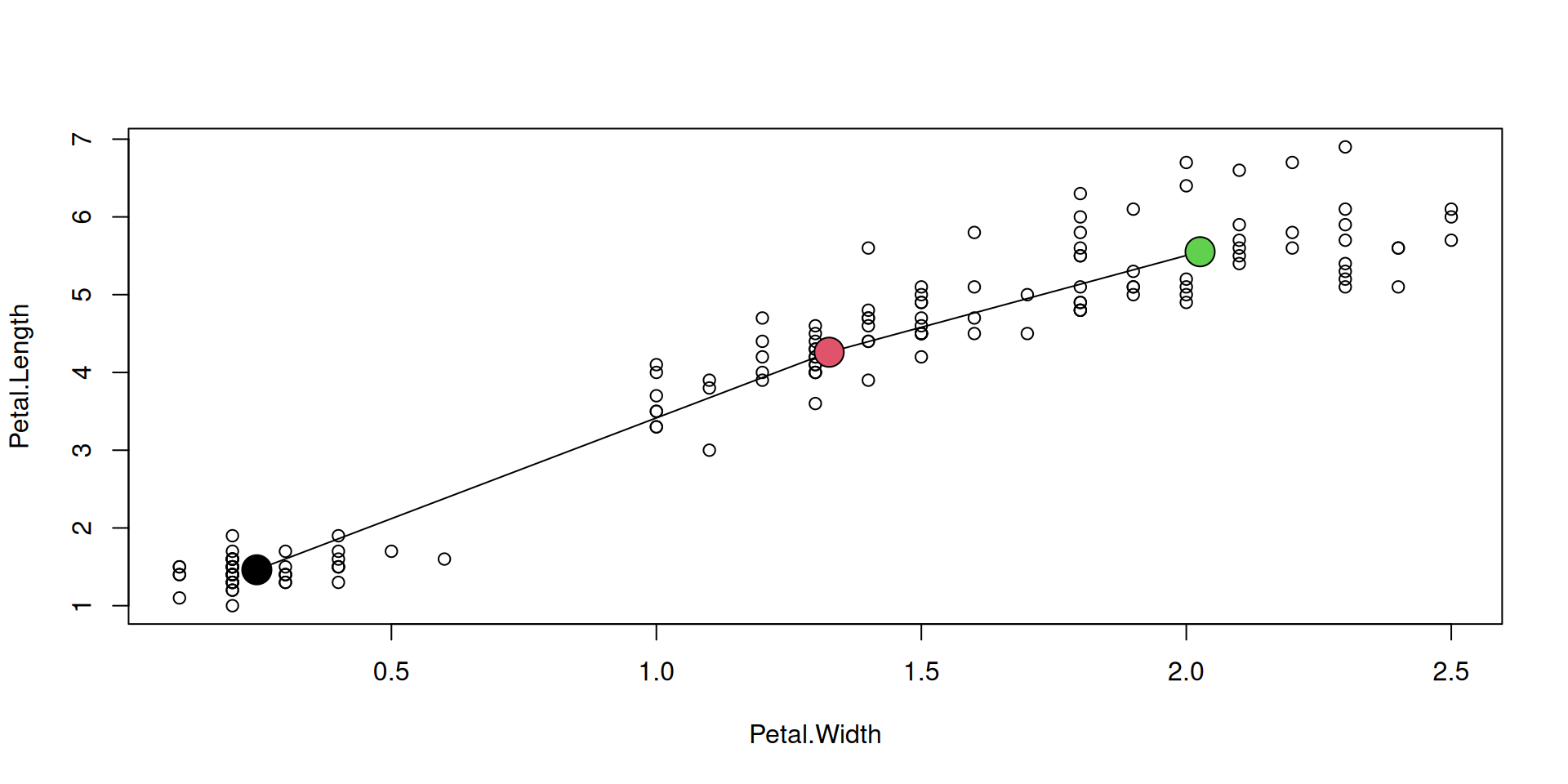
It can make sense to connect some points in a general scatterplot by lines.
The augmenting function lines() can do this.
Here, we want to connect the (x,y) means of our three species:
R Plots (base) – plot()
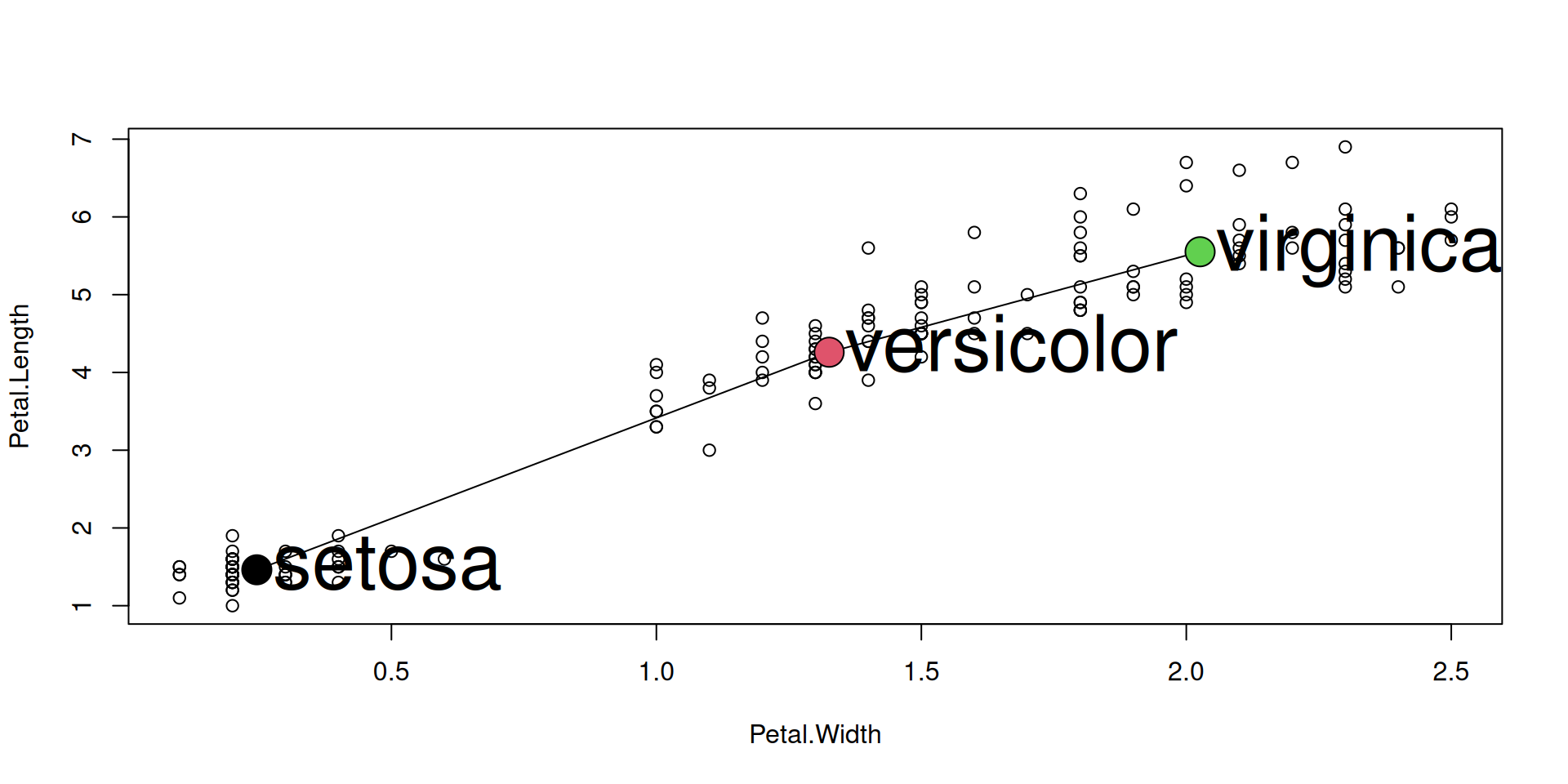
Annotate individual points:
R Plots (base) – plot()
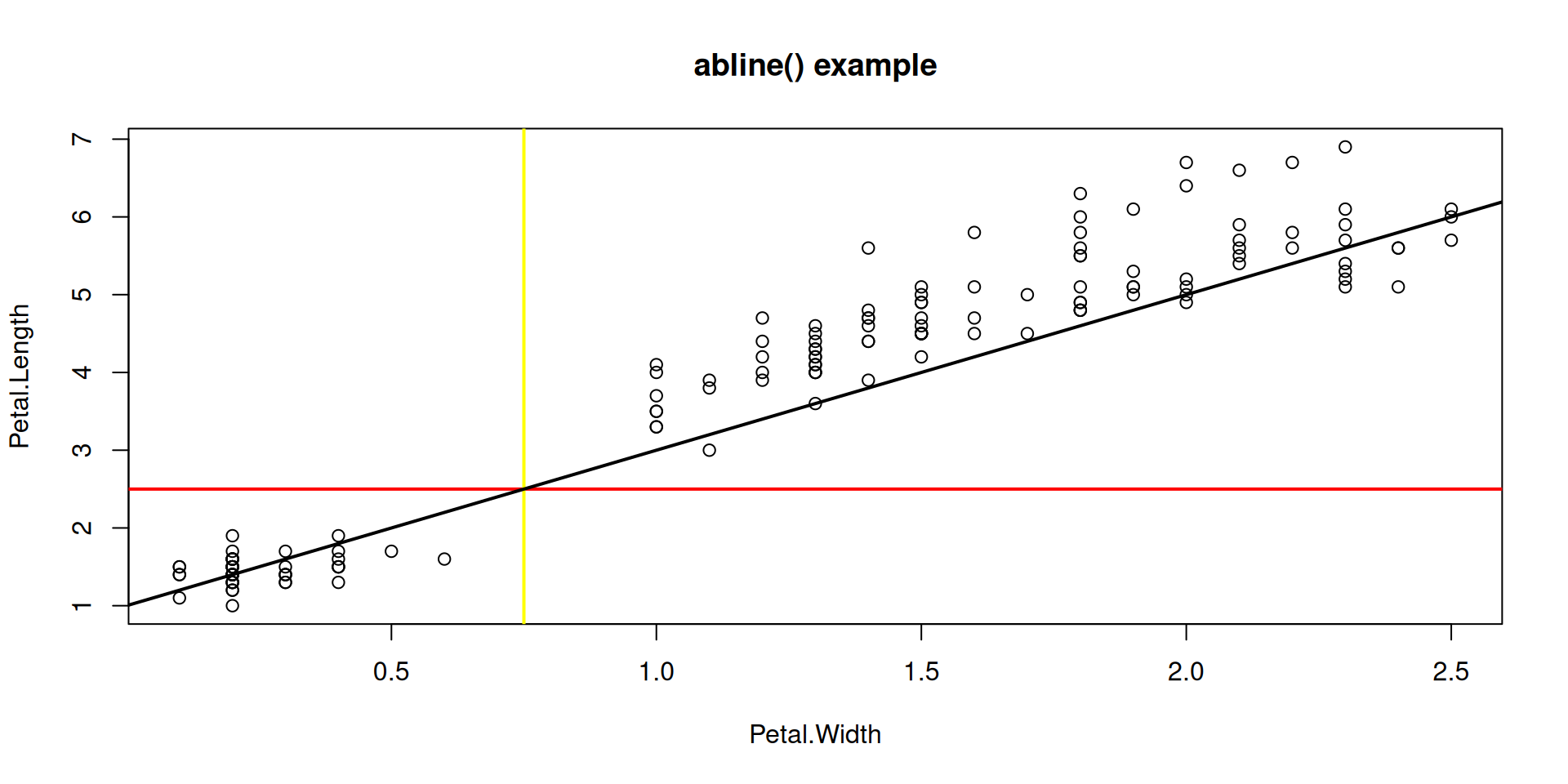
Function abline() adds indicator lines to a plot.
R Plots (base) – plot()
Function abline() adds indicator lines to a plot.
Lines marking locations or slopes of interest:
R Plots (base) – layout()
Several plots can be combined on the same page in a grid-like layout.
The grid is specified by a matrix of possible plot positions, like so:
The first plot will go to grid position 1, the second to position 2 … .
R Plots (base) – layout()
layout(m) ## read the layout matrix
use_cols = species_colors[iris$Species]
## 1
plot(Sepal.Length ~ Sepal.Width, data=iris,
pch=21, col=use_cols, bg=use_cols,
cex.lab=2)
## 2
plot(Petal.Length ~ Petal.Width, data=iris,
pch=21, col=use_cols, bg=use_cols,
cex.lab=2)
## 3
plot(Sepal.Length ~ Petal.Length, data=iris,
pch=21, col=use_cols, bg=use_cols,
cex.lab=2)
## 4
plot(Sepal.Width ~ Petal.Width, data=iris,
pch=21, col=use_cols, bg=use_cols,
cex.lab=2)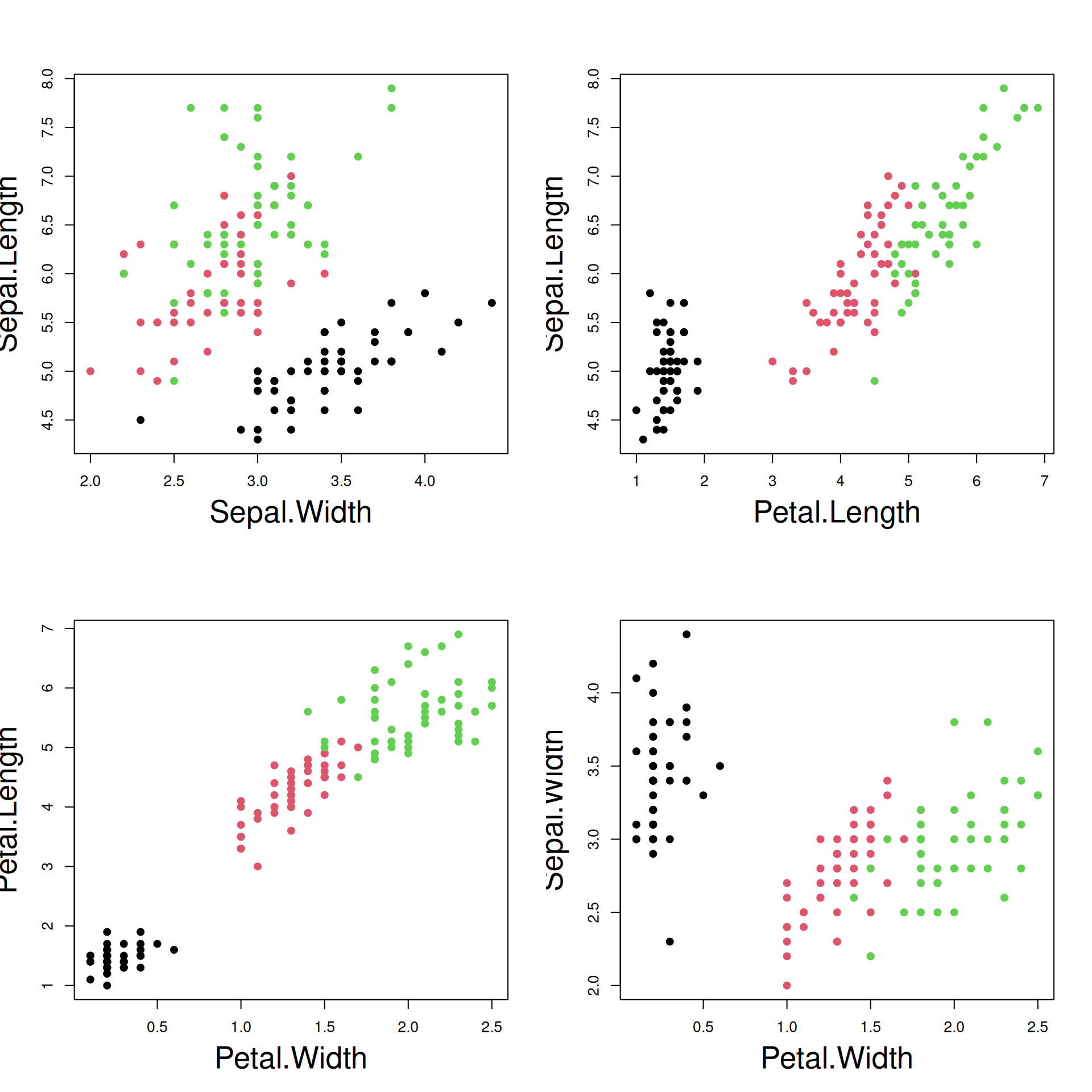
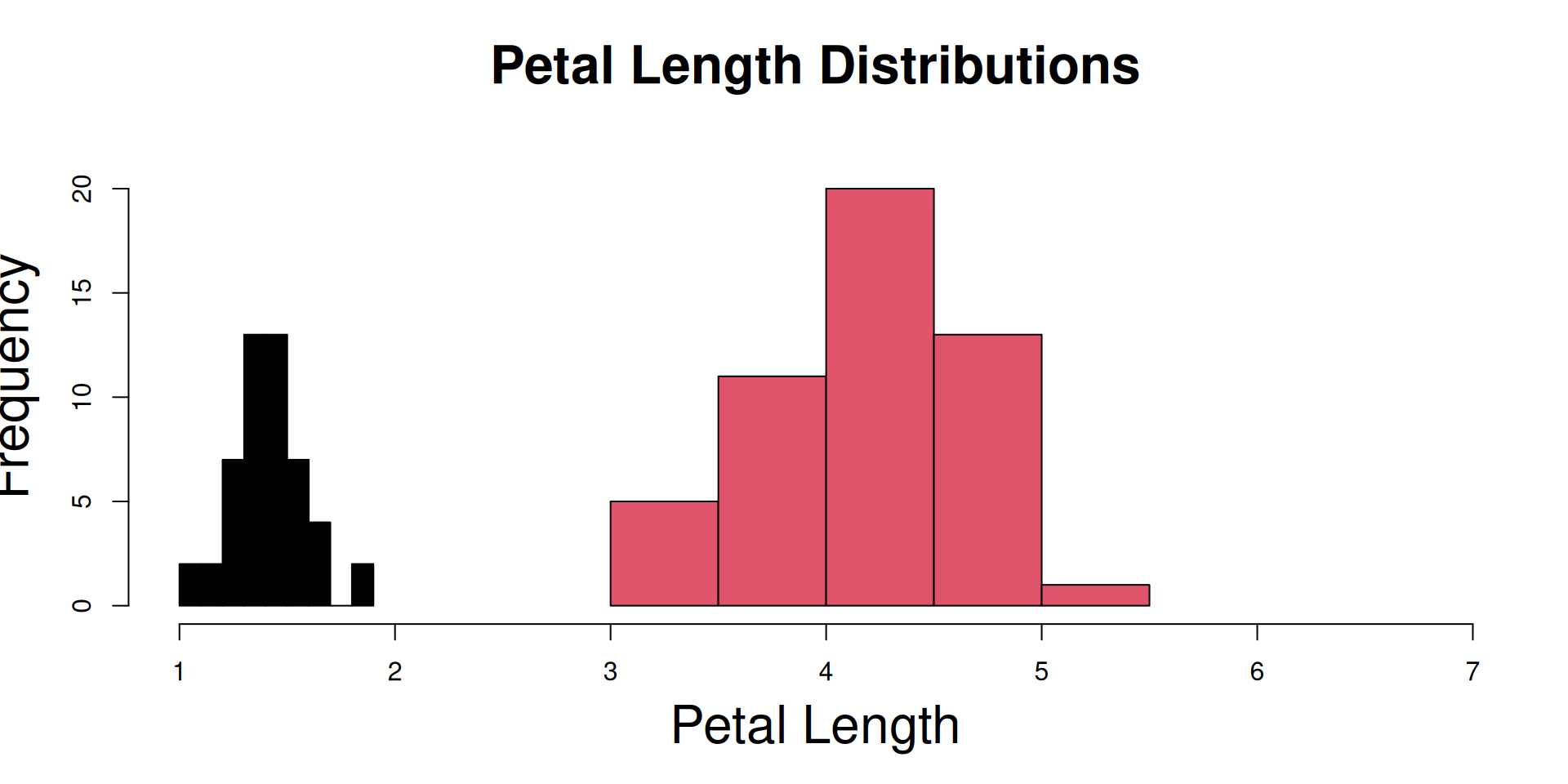
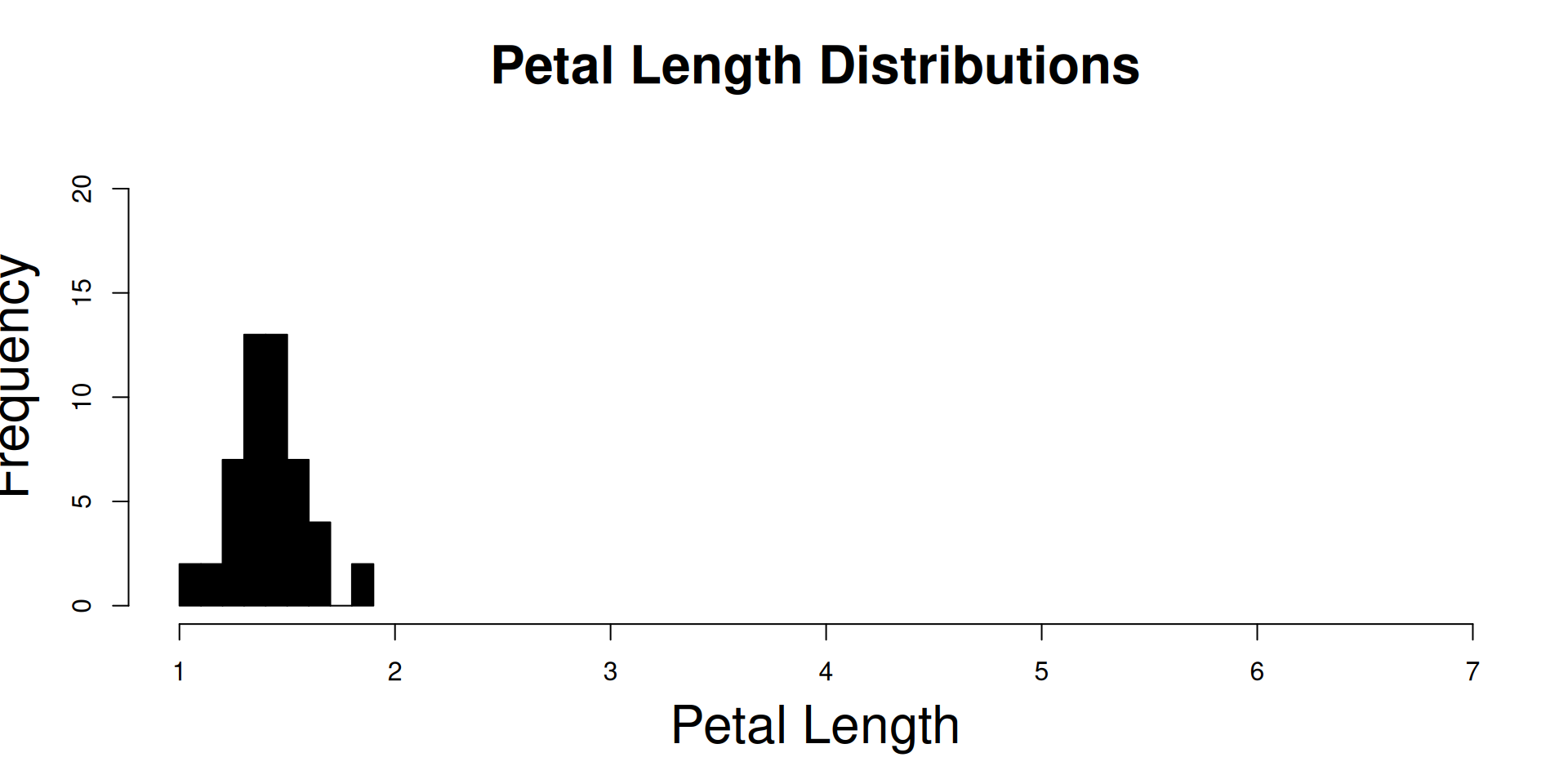
R Plots (base) – hist()
The hist() function is one of those “dual use functions”:
With add=FALSE, it initializes the device and the coordinate system, while
with add=TRUE, its output goes directly to an existing plot.
setosa <- subset(iris,Species=="setosa")
versicolor <- subset(iris,Species=="versicolor")
virginica <- subset(iris,Species=="virginica")
## Plot the histogram of setosa,
## and initialize the entire plot:
hist(setosa$Petal.Length,
col=species_colors["setosa"],
add=FALSE, ## this is the default
## initialize to full x range !
xlim=range(iris$Petal.Length),
## full y range you usually
## only know after some trials ..
ylim=c(0,22),
## x-axis label
xlab="Petal Length",
## larger axis labels:
cex.lab = 2,
main = "Petal Length Distributions",
## larger title:
cex.main = 2
)
R Plots (base) – hist()
The hist() function is one of those “dual use functions”:
With add=FALSE, it initializes the device and the coordinate system, while
with add=TRUE, its output goes directly to an existing plot.
R Plots (base) – hist()
The hist() function is one of those “dual use functions”:
With add=FALSE, it initializes the device and the coordinate system, while
with add=TRUE, its output goes directly to an existing plot.
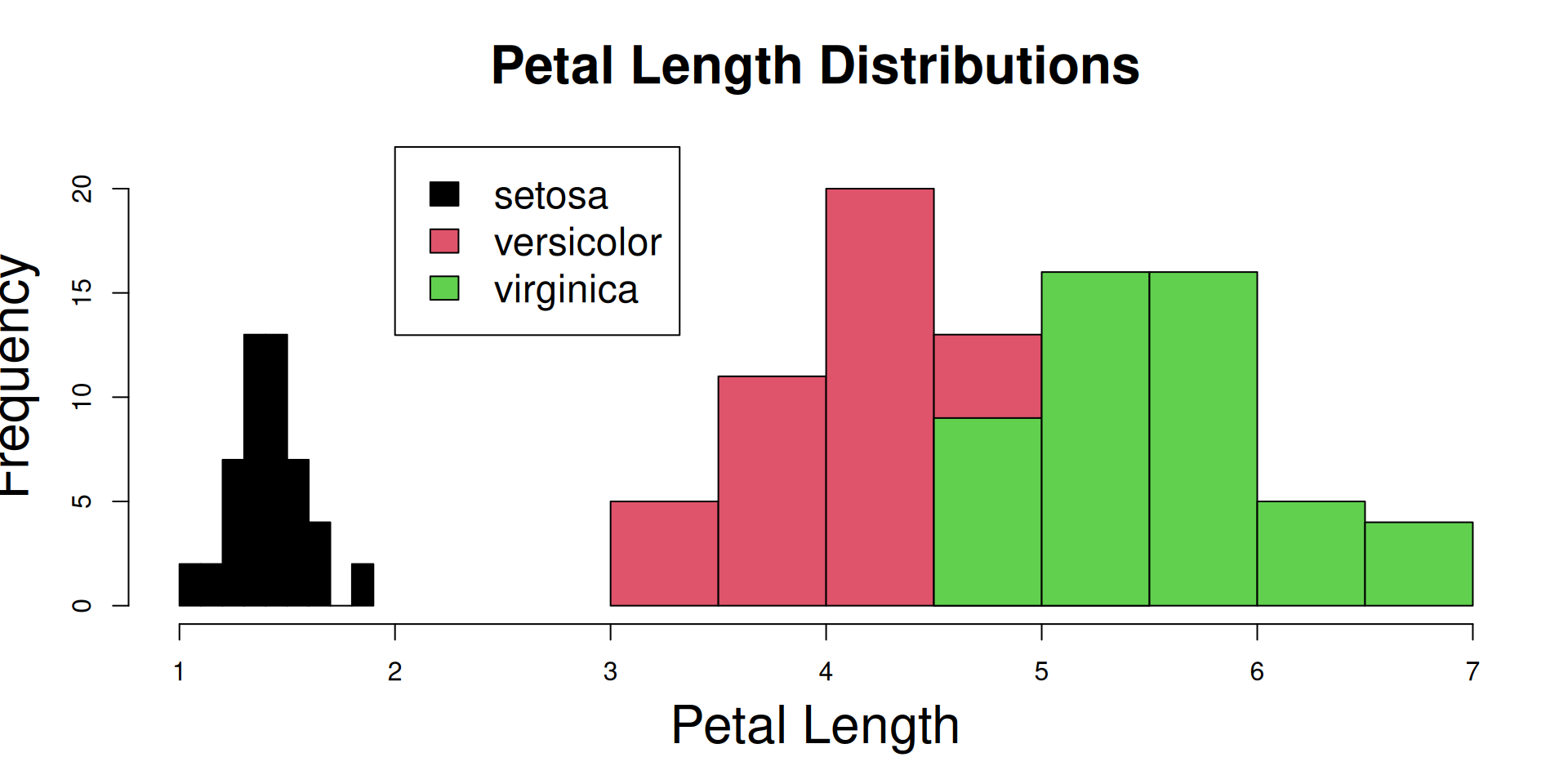
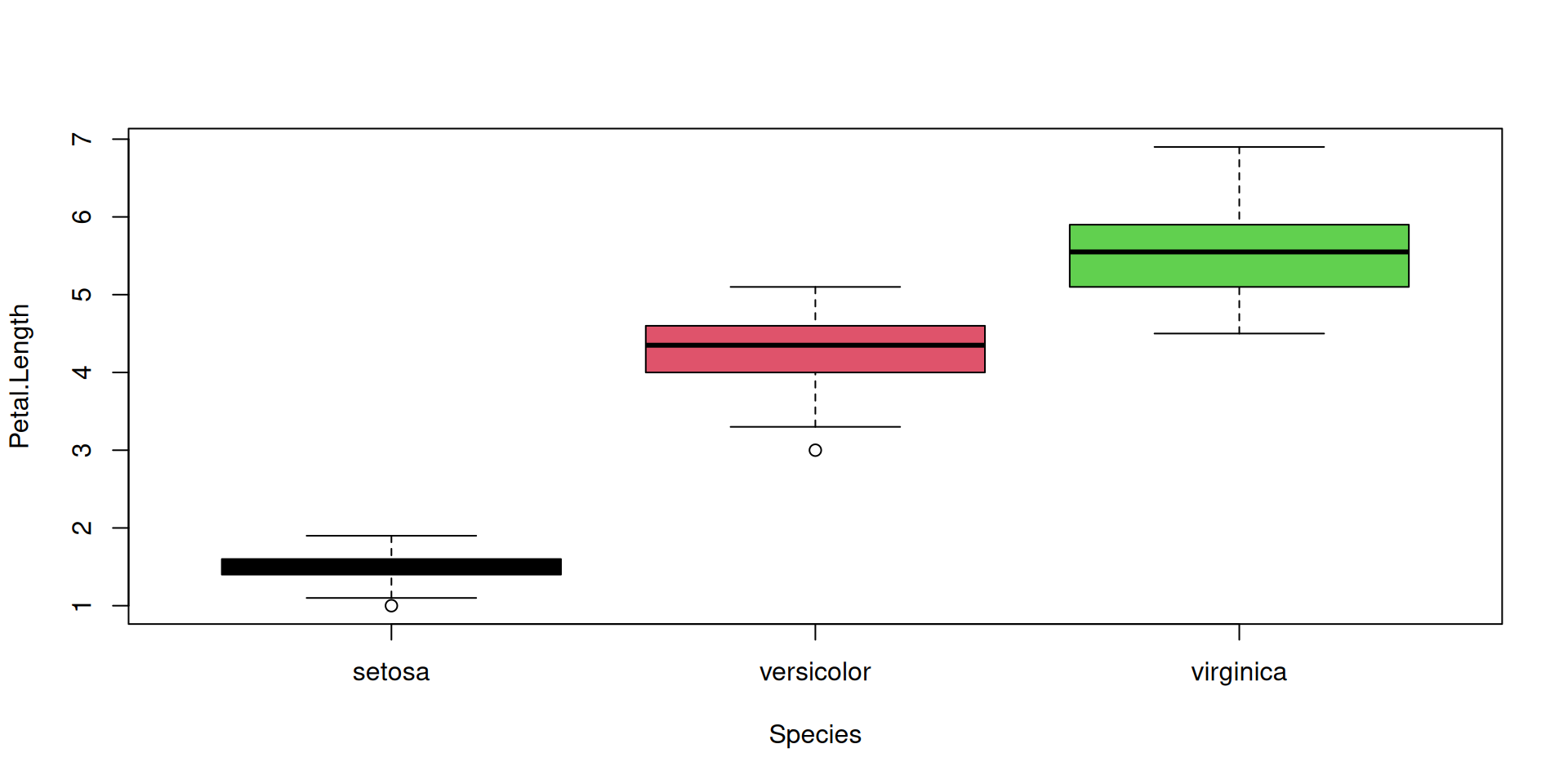
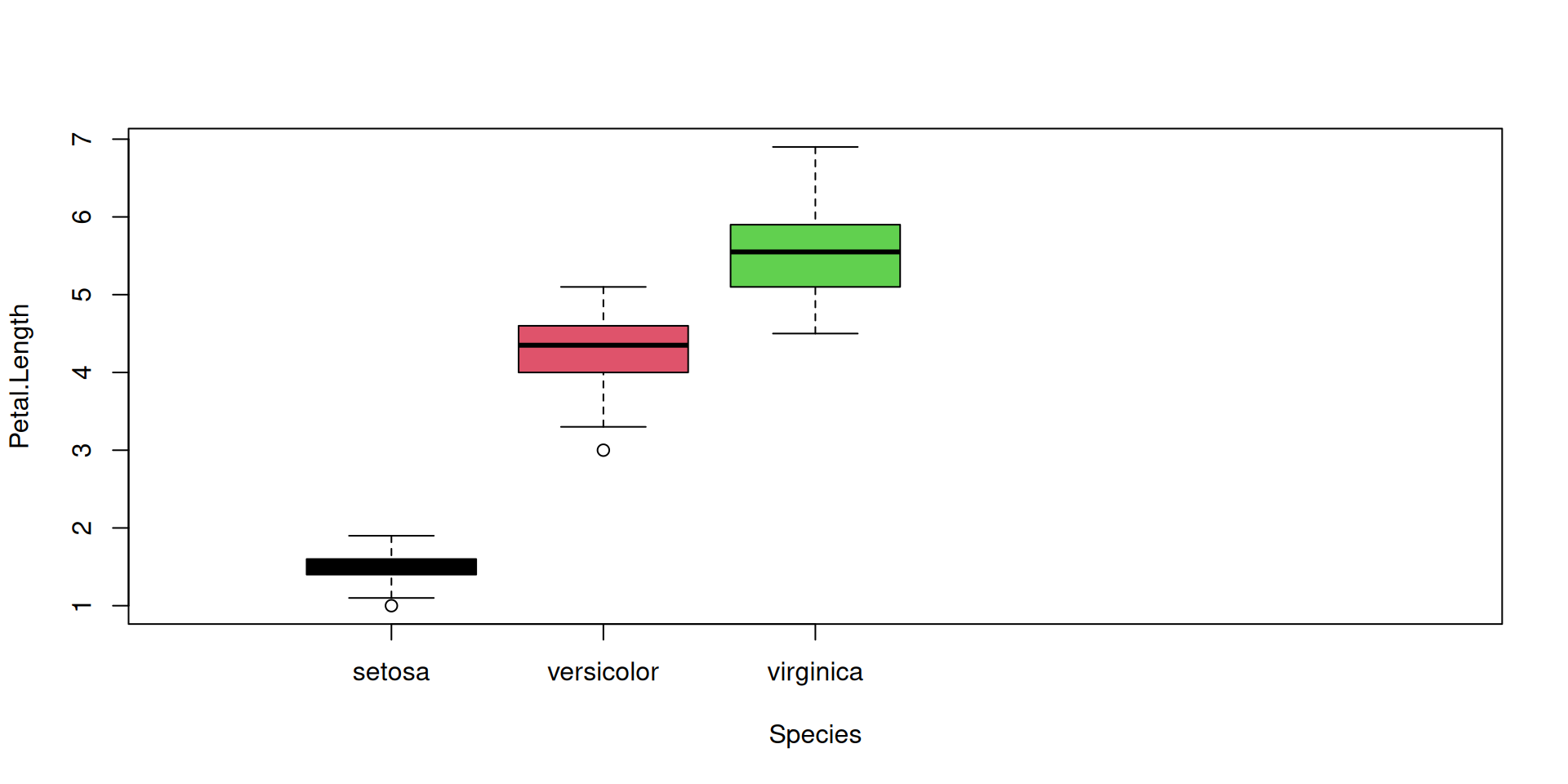
R Plots (base) – boxplot()
The boxplot() function has “dual-use” capabilities, too.
However it can accept a formula with a factor on the right hand side, and it will split the dataset automatically according to the factor levels. So we can plot all species at once:
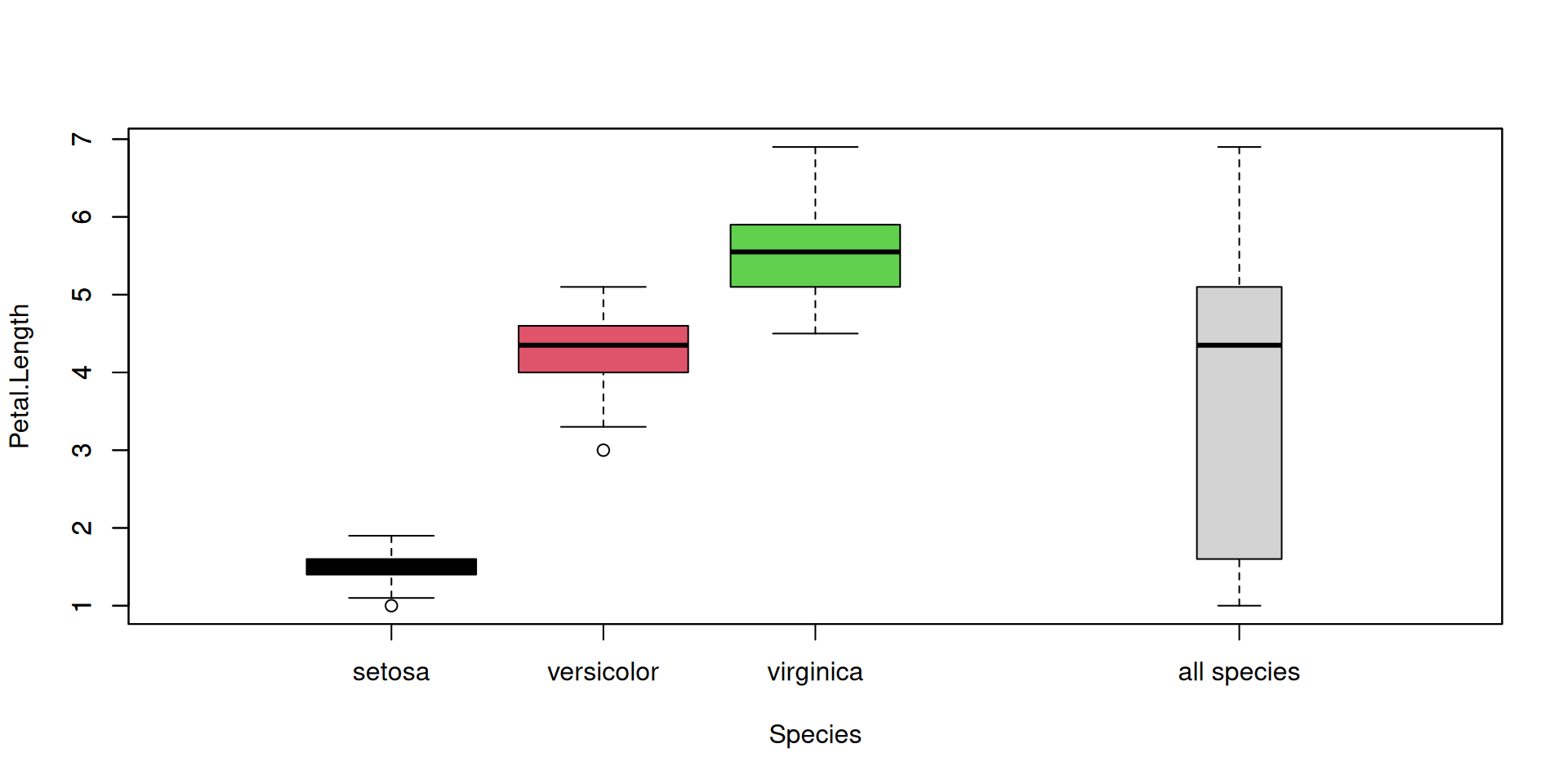
R Plots (base) – boxplot()
Let’s add a boxplot for the global Petal.Length distribution (all species merged):
R Plots (base) – Saving Plots From RStudio
R Plots (base) – Saving Plots From RStudio
Digression: Software is Usually Built From Bits and Pieces
- … they come under the names of subroutines, macros, functions …
- … in code, they are used like ’commands’:
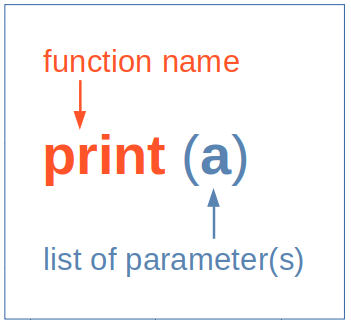
- Actually the function name invokes a piece of hidden code
- … hiding complexity
- … yet allowing to easily access complex algorithms
- … and to make local extensions of a language